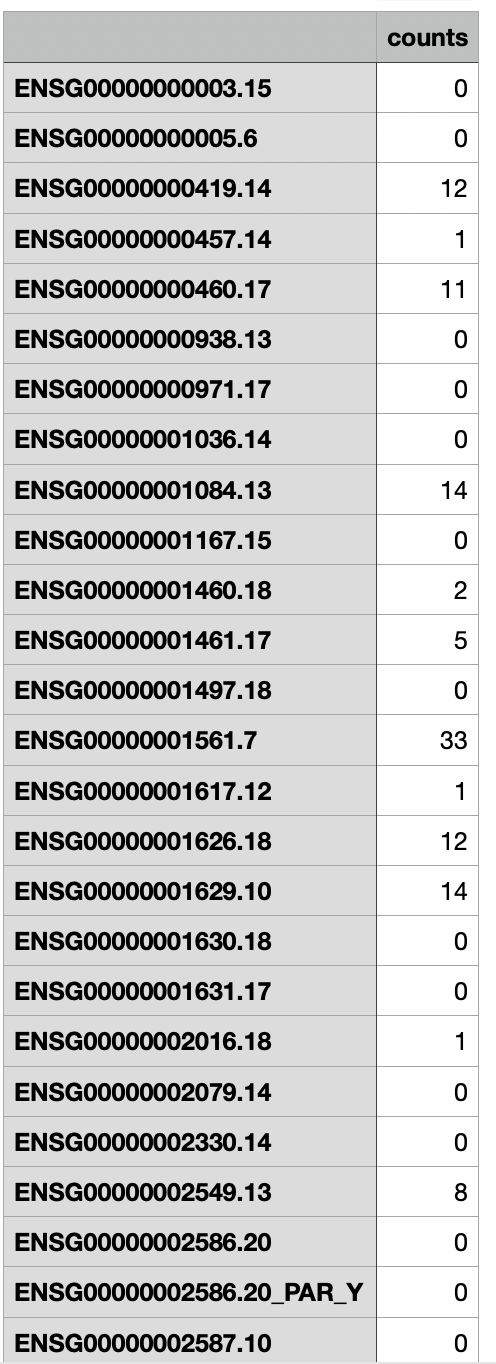
I have 24 samples on which smallRNA seq was performed, most of the genes have 0 counts o0r very low counts. Is this normal for microRNA seq? Attached a screenshot of .csv file with count matrix.
I have three different treatment groups. Due to this, all the treatment groups have same differentially expressed microRNAs but the log2fold change is different. Thank you!
Code should be placed in three backticks as shown below
# include your problematic code here with any corresponding output
# please also include the results of running the following in an R session
sessionInfo( )


Hi @ATpoint,
Thanks for your comments. However, when I perform DESeq2, I get the following results even after filtering for miRNA, not sure if its a DESeq2 problem or not. Only 45 miRNAs have some log2foldchange value in it, the rest do not.
Basemean of zero means these genes have no counts, it's something upstream of DESeq2.
Thanks so much @atpoint. So that means all the miroRNAs should have had some count value? Its unusual to have a countmatrix with such counts for microRNA sequencing?
It's the wrong forum here, please ask somewhere else (biostars or reddit), adding details how samples were made and how the preprocessing were done. Please do not add that here.
Thanks for your response. I have posted it on biostars.