Entering edit mode
I'm wondering which way should I go about comparing modules in multiple networks
I have 3 signed nets with a similar number of modules between 22-27.
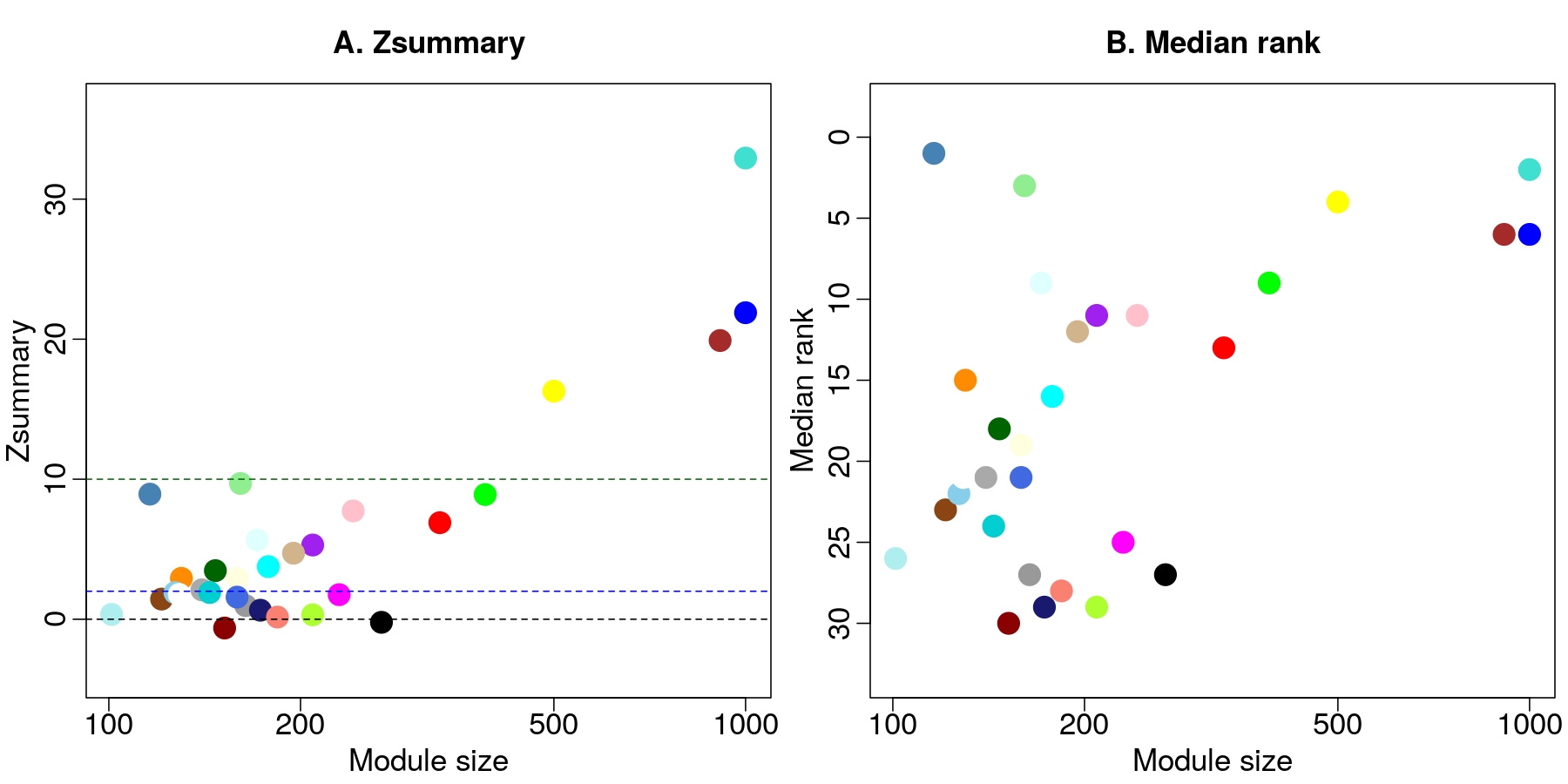
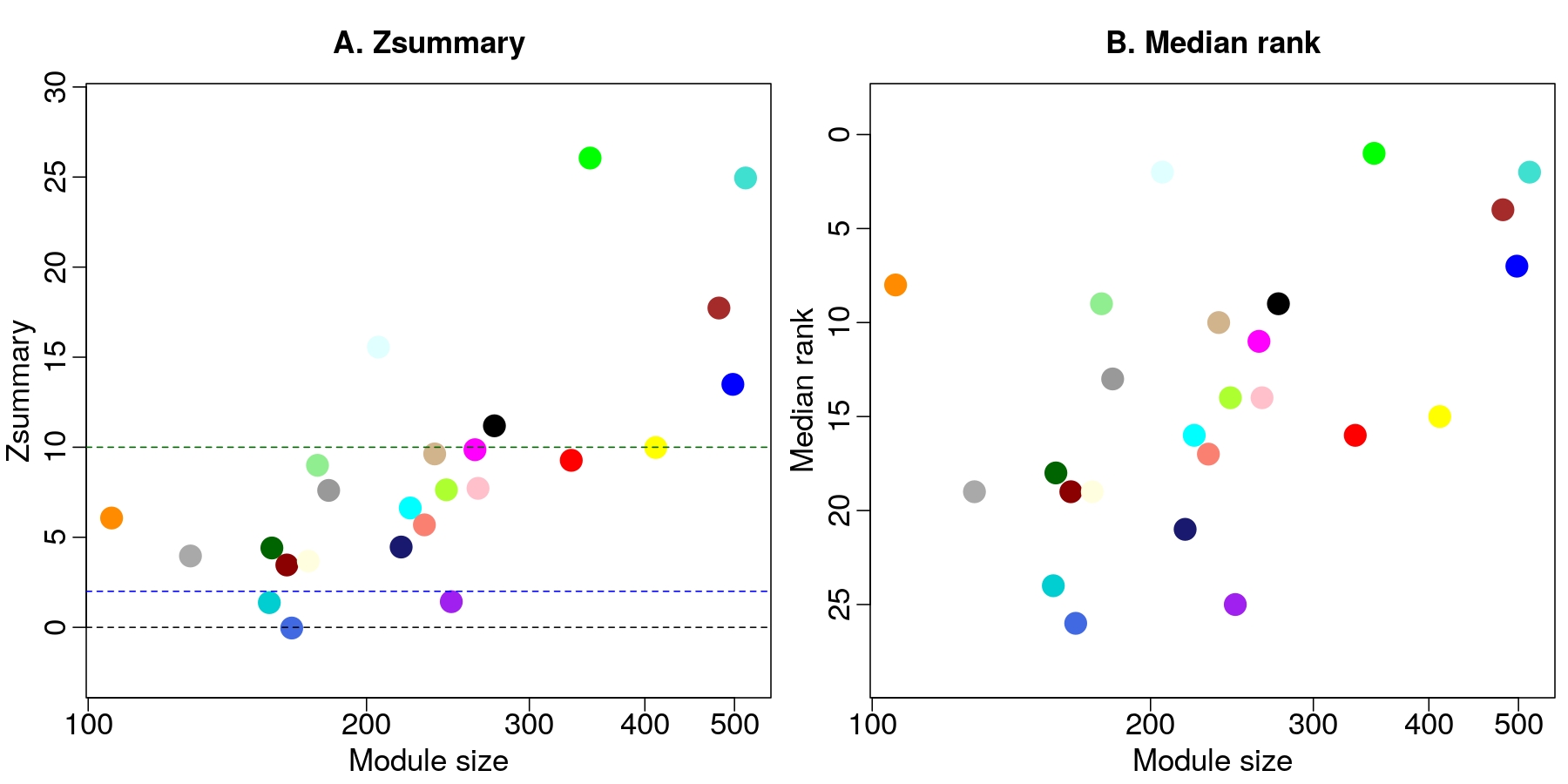
I have done module preservation in
net1 > net2
Now, I would like to see how the expression of genes in net2 as assigned to module colors in net1 is preserved in net 3
I have done net2 > net3
but I believe I cannot say, for example, that those 3 biggest modules well preserved in net1 > net2 are similarly the 3 biggest ones preserved in net2 > net3, bc just looking at it I can see they are half the size.