Hi everyone,
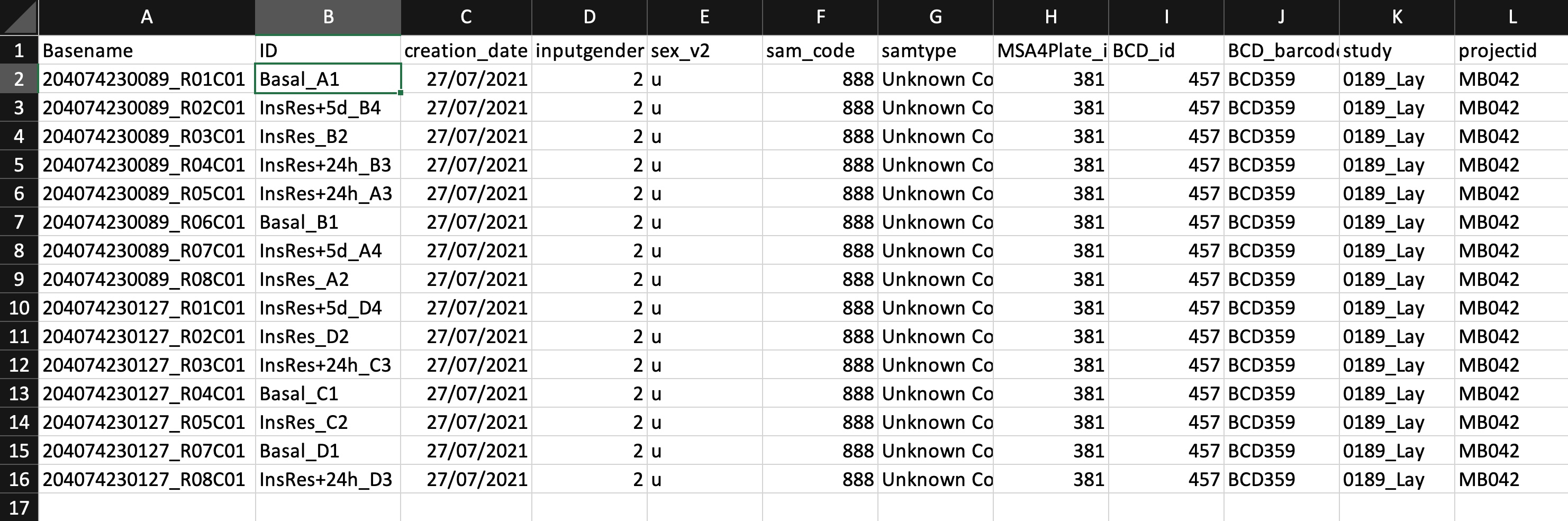
I'm still pretty much an R novice and I'm trying to run an analysis on some EPIC array data I've been given. I am trying to analyse this to understand whether there is differential methylation between 3 different conditions. I have been following the minfi tutorial (https://www.bioconductor.org/help/course-materials/2015/BioC2015/methylation450k.html#quality-control) and trying to adapt the script to make it work with my data. In the tutorial that I'm using, they're analysing how methylation is altered by sex or age, and I need to look at how methylation is altered by insulin resistance. However, I'm getting an error and I'm not sure how to fix it. I think that it's probably related to the sample sheet, or that I'm calling the wrong column. Below is a screenshot of the sample sheet:
I am running the package minfi (version 1.46.0) in RStudio. The R version I have is 4.3.1 and RStudio is 2022.07.1, build 554.
#Load packages
library(minfi)
library(sva)
#Set working directory
setwd("/Users/matthewsinton/Desktop/Manchester Postdoc/EPIC Array Data/IDAT")
#Set location for IDAT files
dataDirectory <- "/Users/matthewsinton/Desktop/Manchester Postdoc/EPIC Array Data/IDAT"
# list the files
list.files(dataDirectory, recursive = TRUE)
#Set location for sample sheet
targets <- read.metharray.sheet(dataDirectory, pattern="SampleSheet.csv")
targets
# read in the raw data from the IDAT files; warnings can be ignored.
RGSet <- read.metharray.exp(targets=targets)
# Get an overview of the data
phenoData <- pData(RGSet)
phenoData[,1:6]
manifest <- getManifest(RGSet)
manifest
head(getProbeInfo(manifest))
#MethylSet and RatioSet. A MethylSet objects contains only the methylated and unmethylated signals
MSet <- preprocessRaw(RGSet)
MSet
head(getMeth(MSet)[,1:3])
head(getUnmeth(MSet)[,1:3])
RSet <- ratioConvert(MSet, what = "both", keepCN = TRUE)
RSet
beta <- getBeta(RSet)
#Genomic Ratio Set. The function mapToGenome applied to a RatioSet object will add genomic coordinates to each probe together with some additional annotation information.
GRset <- mapToGenome(RSet)
GRset
beta <- getBeta(GRset)
M <- getM(GRset)
CN <- getCN(GRset)
sampleNames <- sampleNames(GRset)
probeNames <- featureNames(GRset)
pheno <- pData(GRset)
#To return the probe locations as a GenomicRanges objects, one can use the accessor granges:
gr <- granges(GRset)
head(gr, n= 3)
annotation <- getAnnotation(GRset)
names(annotation)
#Run QC on the data
head(getMeth(MSet)[,1:3])
head(getUnmeth(MSet)[,1:3])
qc <- getQC(MSet)
head(qc)
plotQC(qc)
densityPlot(MSet, sampGroups = phenoData$Sample_Group)
densityBeanPlot(MSet, sampGroups = phenoData$Sample_Group)
controlStripPlot(RGSet, controls="BISULFITE CONVERSION II")
qcReport(RGSet, pdf= "/Users/matthewsinton/Desktop/Manchester Postdoc/EPIC Array Data/qcReport.pdf")
#Preprocessing and normalisation
MSet.illumina <- preprocessIllumina(RGSet, bg.correct = TRUE,
normalize = "controls")
MSet.swan <- preprocessSWAN(RGSet)
GRset.quantile <- preprocessQuantile(RGSet, fixOutliers = TRUE,
removeBadSamples = TRUE, badSampleCutoff = 10.5,
quantileNormalize = TRUE, stratified = TRUE,
mergeManifest = FALSE, sex = NULL)
MSet.noob <- preprocessNoob(RGSet)
GRset.funnorm <- preprocessFunnorm(RGSet)
snps <- getSnpInfo(GRset)
head(snps,10)
GRset <- addSnpInfo(GRset)
GRset <- dropLociWithSnps(GRset, snps=c("SBE","CpG"), maf=0)
#Identifying DMPs and DMRs
beta <- getBeta(GRset.funnorm)
ID <- pData(GRset.funnorm)$ID
dmp <- dmpFinder(beta, pheno = ID , type = "continuous")
head(dmp)
The error I get is:
Error in smooth.spline(lambda, vec.p0, w = ncs.weights, df = 3) :
missing or infinite values in inputs are not allowed
Any help solving this issue would be super helpful. As I say, I'm still a novice with R, so very happy to provide any info I've missed or to clarify anything.


Hi, thanks so much for helping here. Unfortunately this also didn't work, and I got the following error when I ran that bit of script:
Do you have any ideas why that might be?
Thanks!