Entering edit mode
Hi,
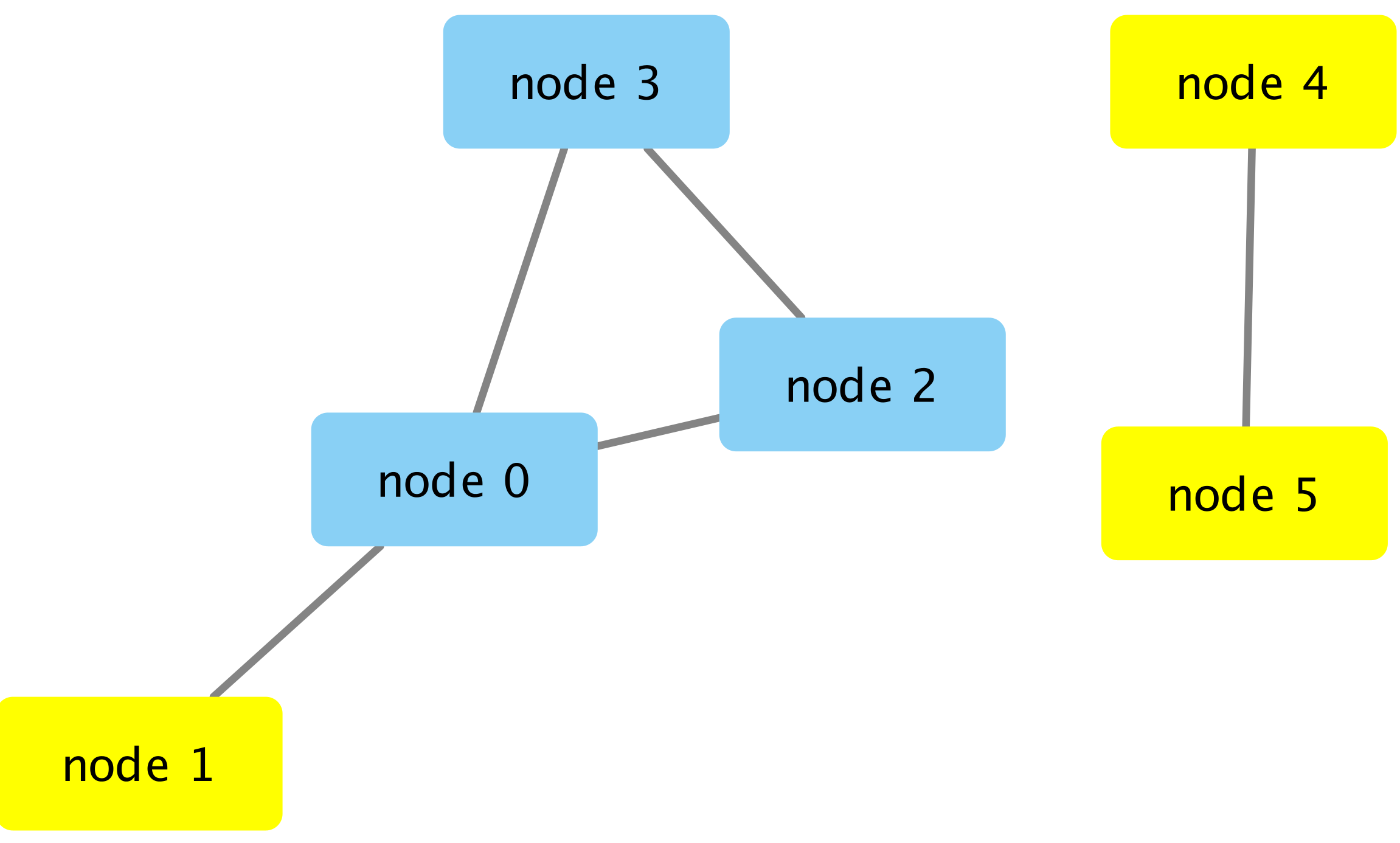
In R-controlled Cytoscape, I would like to select nodes within regions with more than two nodes OR more than one edge. With the code below, nodes 1, 4 and 5 get selected. My goal is to select only nodes 4 and 5 (see image).
Any help would be greatly appreciated. Thanks!
David
library(RCy3)
nodes <- data.frame(
id = c("node 0","node 1","node 2","node 3", "node 4", "node 5"),
stringsAsFactors = FALSE)
edges <- data.frame(
source = c("node 0","node 0","node 0","node 2", "node 4"),
target = c("node 1","node 2","node 3","node 3", "node 5"),
stringsAsFactors = FALSE)
createNetworkFromDataFrames(nodes, edges, title = "test")
createDegreeFilter(
filter.name = 'degree filter',
criterion = c(2,3),
predicate = 'IS_NOT_BETWEEN')