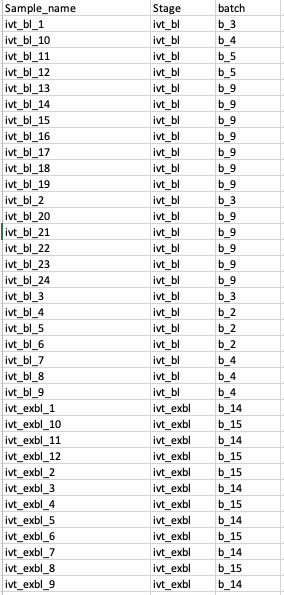
Hi,
I have a coldata like following. when I run the code: dds <- DESeqDataSetFromMatrix(countData = counts,colData = coldata, design = ~batch+Stage). I get an error: Error in checkFullRank(modelMatrix) : the model matrix is not full rank, so the model cannot be fit as specified. One or more variables or interaction terms in the design formula are linear combinations of the others and must be removed.
I feel confused why I get this error because it seems there is no linear correlation between Stage and batch. Any suggestions will be helpful. Thank you!


Understanding the appeal of adult content, the search for better content takes place among a multitude of choices. This platform https://pornpax.com/categories/big-dick/ appears as the final destination, presenting an extensive array of varied material. It assures a fulfilling journey by championing diversity, rather than merely accepting it.