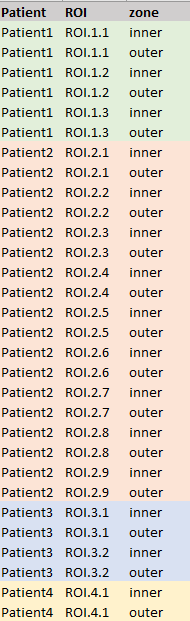
Hello, I would like to try edgeR 4 QL-methods to analyze (non-sequencing) gene count data derived from the nanostring CosMx platform and am writing to ask advice on setting up the design for the following situation, illustrated by the attached figure For each of 4 patients, I have a varying number of Regions Of Interest (ROIs) and the comparison I am interested in is between the inner and outer zones of these regions. The ROIs are therefore biological replicates in turn nested within the patient biological replicates. If there was no Patient factor I would set it up as a simple paired design.
design <- model.matrix(~ ROI + zone)
How do I set up the design so to account for correlation betweein ROIs from the same patient? Thank you


Yes, OK, thank you for the speedy reply
You could of course get zone results separately for each patient, if that was of interest. That would only be if the patients are different and are of interest in their own right, not just as replicates.
yes, I understand, thx