Entering edit mode
I have RNA-Seq data that is collected from different centers and I want to analyze it using ReactomeGSA-Camera but I am not sure how to adjust for batch effect and unwanted variation on the web tool. I already performed RUVg correction for that purpose and I have a matrix of estimated factors of unwanted variation. However, when I insert these factors in the sample annotation on ReactomeGSA, it gives me this error.

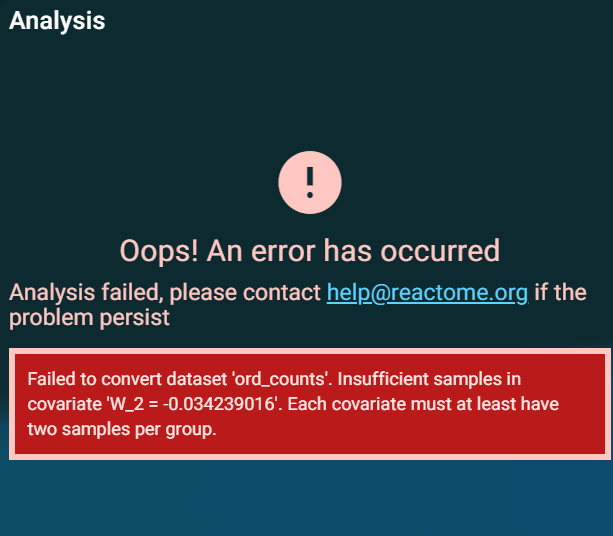

Hi! The factors need to be categorical, i.e. the ID / name of the center.