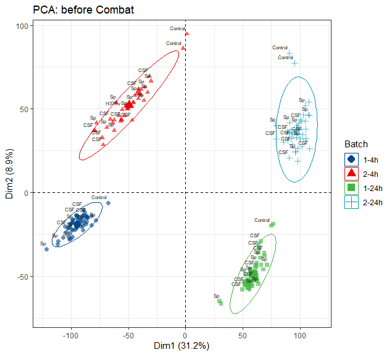
Hello, I am working with RNAseq data which was acquired from experiments as following:
- PBMCs were extracted from blood of healthy donors, then extracting monocytes, pooling monocytes of different healthy donors, then differentiate pooled monocytes into macrophages.
- After that, Mycobacterium tuberculosis isolates were infected into macrophages, I also included lab strain H37Rv, and no-infection group.
- Then macrophages were collected RNA at 4h and 24h post-infection.
- Because the heavy workload, I divided into 2 batches, each batch included 2 samples of no-infection group, 2 samples of H37Rv as control.
- Macrophages from 2 batches still from the same heathy donors.
- After collecting all RNA samples, RNA was sequenced at the same time.
When I did PCA, there was big difference between batches, even for no-infection and H37Rv group as picture below
I would like to ask that there are any method for batch effect removal or normalization to combine the data between batches.
I was thinking to normalize Mtb strain to no-infection group in each batch before comparing between isolates.
Thank you in advance.