Hi, I am trying to learn about DEG and Deseq2 and using the Beginner's guide
1) does running dds <- DESeq(dds) automatically apply "Normal" shrinkage and also apply rlog ?
2) in the beginners guide section5, it says to transform using rld <- rlog( dds ) .. is this just showing what dds has already done? Or does this need doing each time?
3) I attempted to perform deseq2 on my data - on my volcano plot, gene PALB2 has a strong pval, but only modest LFC, and on the volcano plot it only shows small/moderate expression. how can this be explained?
many thanks

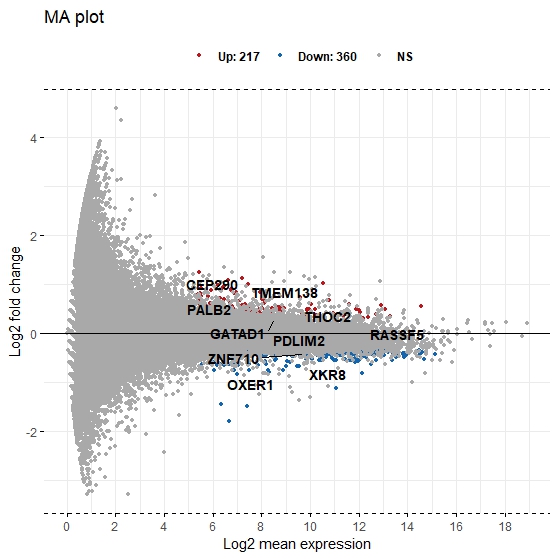
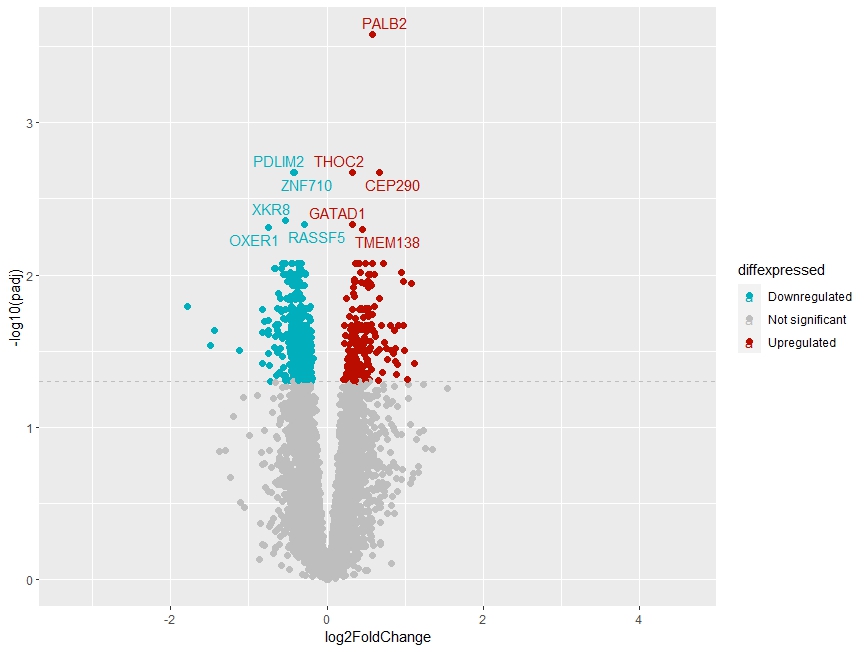

Cross-posted to Biostars: https://www.biostars.org/p/9602828/