When using the linked Pr2 V5 database with DECIPHER IDtaxa, I have found that there are no ranks associated with the taxonomic assignments given by the classifier. This has resulted in my inability to form a taxonomy table based on the assignments given by the classifier. When I run the code below, I get a taxonomy table that is entirely NA values. I'm not sure how to solve this, and if there is a fix that someone knows to include ranks in the classifier, I would be highly appreciative.
#assign taxonomy using IDtaxa and Pr2 DB
dna.20 <- DNAStringSet(getSequences(seqtab.2020))
file_trained = "/Users/rettigm/Downloads/pr2_version_5.0.0_SSU.decipher.trained.rds"
trainingSet <- readRDS(file_trained)
ids.20 <- IdTaxa(dna.20, trainingSet, strand = "top", processors = NULL, verbose = TRUE)
ranks.pr2 <- c("kingdom", "division", "phylum", "class", "order", "family", "genus", "species")
taxid.20 <- t(sapply(ids.20, function(x) {
m <- match(ranks.pr2, x$rank)
taxa.20 <- x$taxon[m]
taxa.20[startsWith(taxa.20, "unclassified_")] <- NA
taxa.20
}))
colnames(taxid.20) <- ranks.pr2; rownames(taxid.20) <- getSequences(seqtab.2020)
sessionInfo( ): R version 4.4.0 (2024-04-24) Platform: x86_64-apple-darwin20 Running under: macOS Ventura 13.2.1
This is the output of each row of the classifier, which has no rank associated with it, an issue I do not experience with the SILVA database. I am suspecting that this is the source of issue I'm having attempting to build the taxonomy table.
This is the issue I'm having with the taxonomy table, I suspect the lack of rank in the assignment prevents the taxonomy from mapping properly into the taxonomy table.

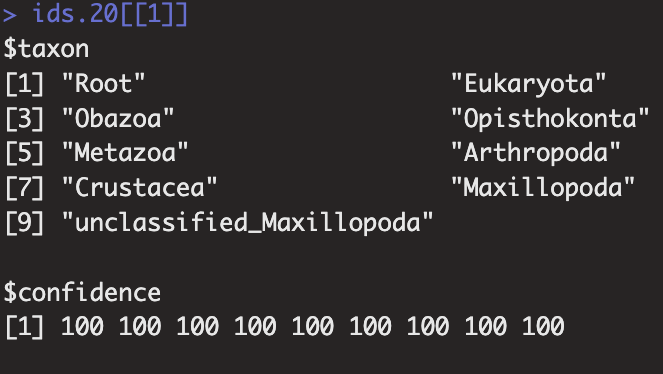
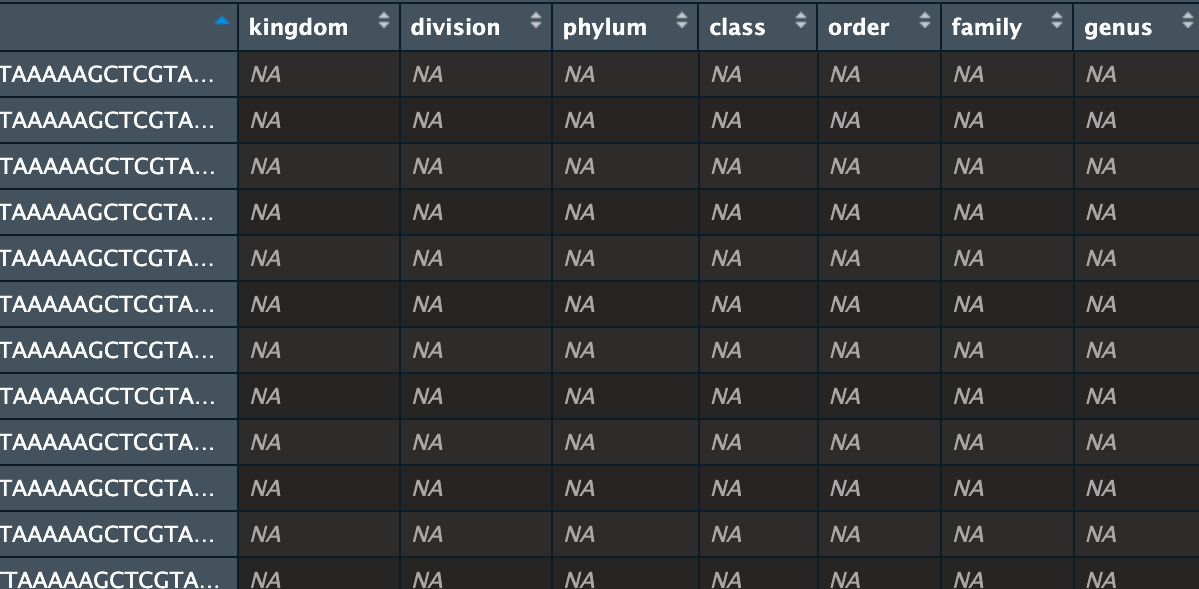

IDTAXA classification can be performed from training sets with or without assigned ranks. In this case it looks like
LearnTaxa()was not given rank information.The pre-trained PR2 classifier available here contains ranks, but the PR2 instructions here do not supply ranks as input into
LearnTaxa().I believe PR2 is a fixed (8 level) taxonomy, so it should be straightforward to add ranks during training. Please reach out if you need help with the training step.
Two things:
PR2 version 5.0 and above have 9 levels and not 8 (up to version 4.14). So the code should be:
PR2 does not use a taxon table but a taxonomy table. So there are no ranks nor taxon_id in the format that is required by
LearnTaxafunction (parameterrank =). I provide a tutorial to get back the assignment table using the files provided by PR2.However the next version of PR2 (5.1) will update the tutorial and the DECIPHER trained set to include a taxid.