Dear all,
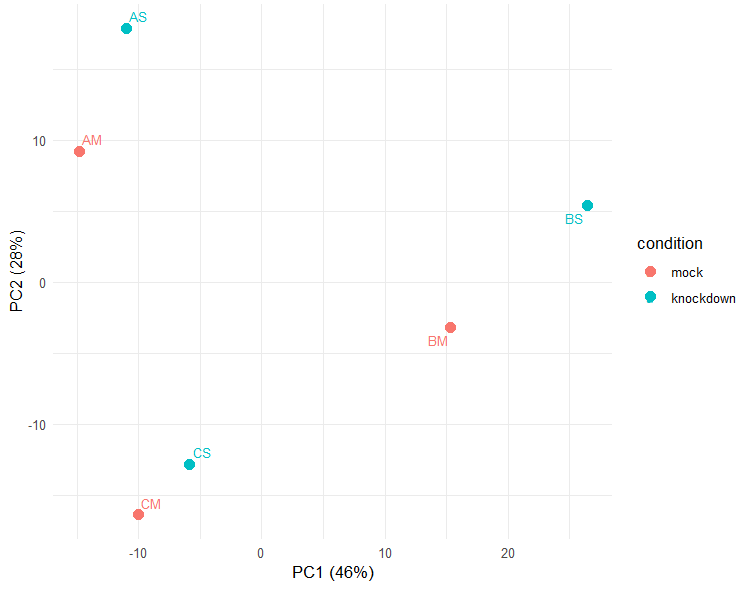
I have a question regarding the consideration of the biological replicates inside the design formula of DESeq2. We performed 2 different approaches, one being (design=~condition) and the other (design= ~replicate + condition) (see Metadata in picture 1) and found out that there were significant variations in number and padj value of DEG detected.
Since PCA plot shows similar tendencies between conditions but also variance between samples from the same group, we were wondering if it would be more accurate to adjust the design to ignore the inherent variance of independent biological replicates and batch effects to maximize differences that are based on our condition of interest.
To put into perspective, first approach gives us 44 DEG (filtered by padj<0,05 only) and the second one 1640 (padj< 0,05), with the second one allowing us to introduce stricter filtering criteria such as |log2foldChange| >1.
Thanks in advance for your help
Christian
PhDStudent in Health Sciences
```