I have a 10x 5' scRNA-seq dataset where Cell Ranger (v9.0.0) reports 64,610 non-empty droplets. This is plausible since it comes from an hashtagged and superloaded GEM-X capture.
I always run DropletUtils::emptyDrops() by way of comparison, and here got very different results: 27,739 non-empty droplets.
Knowing that the default value of the alpha = Inf parameter changed recently-ish, I repeated this with the former default alpha = NULL and recovered numbers comparable to Cell Ranger: 72,162 non-empty droplets.
I'm trying to understand which approach to put more weight on, and conducted some experiments (copied below).
I can share the count matrix, (x; ~300 MB) if it would be helpful.
Any suggestions and explanations appreciated.
Thanks, Pete
library(DropletUtils)
x <- readRDS("x.rds")
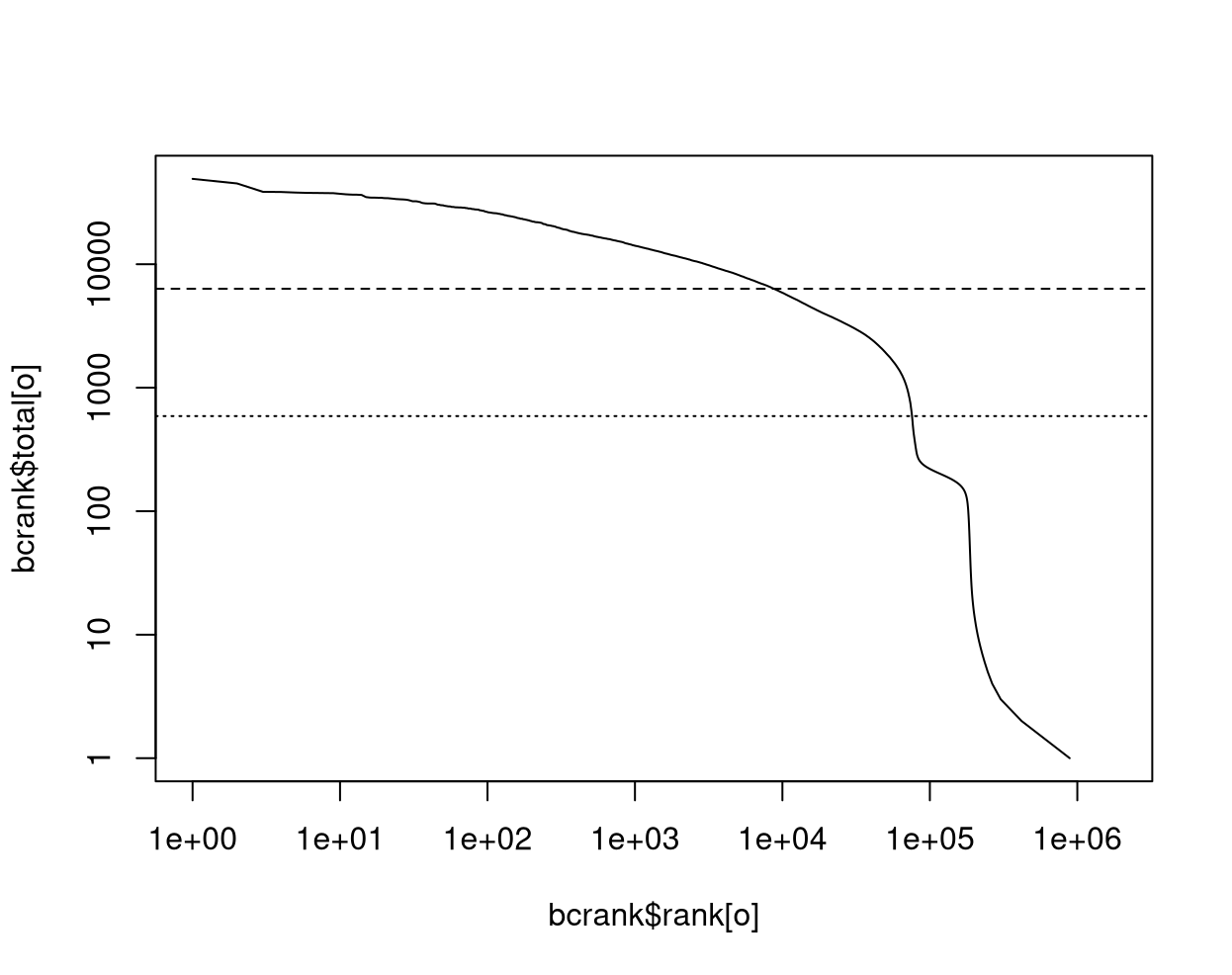
bcrank <- barcodeRanks(x)
uniq <- !duplicated(bcrank$rank)
plot(
bcrank$rank[uniq],
bcrank$total[uniq],
log = "xy",
xlab = "Rank",
ylab = "Total UMI count",
cex.lab = 1.2)
#> Warning in xy.coords(x, y, xlabel, ylabel, log): 1 y value <= 0 omitted from
#> logarithmic plot
abline(h = metadata(bcrank)$inflection, col = "darkgreen", lty = 2)
abline(h = metadata(bcrank)$knee, col = "dodgerblue", lty = 2)
legend(
"bottomleft",
legend = c("Inflection", "Knee"),
col = c("darkgreen", "dodgerblue"),
lty = 2,
cex = 1.2)

# emptyDrops under default settings --------------------------------------------
lower <- 100
set.seed(666)
ed_alpha_inf <- emptyDrops(x, lower = lower, test.ambient = TRUE)
ed_alpha_null <- emptyDrops(x, lower = lower, test.ambient = TRUE, alpha = NULL)
sum(ed_alpha_inf$FDR <= 0.001, na.rm = TRUE)
#> [1] 27739
sum(ed_alpha_null$FDR <= 0.001, na.rm = TRUE)
#> [1] 72162
par(mfrow = c(1, 2))
hist(
ed_alpha_inf$PValue[ed_alpha_inf$Total <= lower & ed_alpha_inf$Total > 0],
xlab = "P-value",
main = "alpha = Inf",
col = "grey80")
hist(
ed_alpha_null$PValue[ed_alpha_null$Total <= lower & ed_alpha_null$Total > 0],
xlab = "P-value",
main = "alpha = NULL",
col = "grey80")

# emptyDrops under expectation of ~60,000 non-empty droplets -------------------
by.rank <- 70000
set.seed(666)
ed_alpha_inf <- emptyDrops(x, by.rank = by.rank, test.ambient = TRUE)
ed_alpha_null <- emptyDrops(x, by.rank = by.rank, test.ambient = TRUE, alpha = NULL)
sum(ed_alpha_inf$FDR <= 0.001, na.rm = TRUE)
#> [1] 30369
sum(ed_alpha_null$FDR <= 0.001, na.rm = TRUE)
#> [1] 49461
# NOTE: Extract lower (doesn't depend on alpha)
lower <- metadata(ed_alpha_inf)$lower
lower
#> [1] 992
par(mfrow = c(1, 2))
hist(
ed_alpha_inf$PValue[
ed_alpha_inf$Total <= lower & ed_alpha_inf$Total > 0],
xlab = "P-value",
main = "alpha = Inf",
col = "grey80")
hist(
ed_alpha_null$PValue[
ed_alpha_null$Total <= lower & ed_alpha_null$Total > 0],
xlab = "P-value",
main = "alpha = NULL",
col = "grey80")

# More extreme version of the above --------------------------------------------
by.rank <- 100000
set.seed(666)
ed_alpha_inf <- emptyDrops(x, by.rank = by.rank, test.ambient = TRUE)
ed_alpha_null <- emptyDrops(x, by.rank = by.rank, test.ambient = TRUE, alpha = NULL)
sum(ed_alpha_inf$FDR <= 0.001, na.rm = TRUE)
#> [1] 36717
sum(ed_alpha_null$FDR <= 0.001, na.rm = TRUE)
#> [1] 65752
# NOTE: Extract lower (doesn't depend on alpha)
lower <- metadata(ed_alpha_inf)$lower
lower
#> [1] 220
par(mfrow = c(1, 2))
hist(
ed_alpha_inf$PValue[
ed_alpha_inf$Total <= lower & ed_alpha_inf$Total > 0],
xlab = "P-value",
main = "alpha = Inf",
col = "grey80")
hist(
ed_alpha_null$PValue[
ed_alpha_null$Total <= lower & ed_alpha_null$Total > 0],
xlab = "P-value",
main = "alpha = NULL",
col = "grey80")

# emptyDropsCellRanger() -------------------------------------------------------
# NOTE: Unsure how closely this actually mimics what Cell Ranger is doing, but
# including because it's easy enough to do.
ed_cr <- emptyDropsCellRanger(x, n.expected.cells = 60000)
sum(ed_cr$FDR <= 0.001, na.rm = TRUE)
#> [1] 63190





Session info