My data set contains three factors i would like to analyze in a time-series using the DESeq2 package
I have stimulated vs. unstimulated cells i have knockout vs. wildtype i have three time-points 0,6,24 hours.
I would like to know if it is possible to create an interaction with three terms cell_line:time_point:treatment.
my code would then be:
coldata$time_point <- factor(coldata$time_point)
coldata$treatment <- factor(coldata$treatment)
coldata$cell_line <- factor(coldata$cell_line)
dds_WT_KO <- DESeqDataSetFromTximport(txi = txi.kallisto,
colData = coldata,
design = ~ cell_line + time_point + treatment +
cell_line:time_point +
cell_line:treatment +
time_point:treatment +
cell_line:time_point:treatment)
dds_WT_KO$treatment <- relevel(dds_WT_KO$treatment, ref = "none") # Set "none" as the reference treatment
dds_WT_KO$cell_line <- relevel(dds_WT_KO$cell_line, ref = "WT") # Set "none" as the reference treatment
dds_WT_KO <- DESeq(dds_WT_KO, test="LRT",
reduced = ~ cell_line + time_point + treatment +
cell_line:time_point +cell_line:treatment +
time_point:treatment) # missing the triple interaction term
res_triple_interaction <- results(dds_WT_KO) # Not really sure if this makes sense and how to interpret the results here.
# Extract other specific contrasts
## genotype - Which genes are generally higher/lower in the KO, regardless of time or treatment?
res_genotype <- results(dds_WT_KO, name="cell_line_Il4i1ko_vs_WT")
## treatment -
res_treatment <- results(dds_WT_KO, contrast=list("treatment_CpG_vs_none"))
#Stimulation - Does the KO blunt or enhance the CpG response?
res_ko_effect_on_cpg_response <- results(dds_WT_KO, name="cell_lineIl4i1ko.treatmentCpG")
#Which genes exhibit a time-dependent response to CpG that is significantly different in the Il4i1 KO compared to the WT?
res_time-dependent_cpg_response_6h <- results(dds_WT_KO, name="cell_lineIl4i1ko.time_point6.treatmentCpG")
res_time-dependent_cpg_response_24h <- results(dds_WT_KO, name="cell_lineIl4i1ko.time_point24.treatmentCpG")
I put the questions to the different results I can extract from the resultnames(dds) in the comments in the code.
My main concern is if this kind of triple interaction even possible or makes sense at all? My second quesion is if my interpretation of the contrasts i made are correct here.
thanks in advance
my metadata can be extracted from this table
new("DFrame", rownames = c("Il4i1ko_0h_S22", "Il4i1ko_0h_S21",
"Il4i1ko_0h_S20", "WT_0h_S7", "WT_0h_S6", "WT_0h_S5", "WT0h_1",
"WT0h_2", "WT0h_3", "Il4i1ko0h_16", "Il4i1ko0h_17", "Il4i1ko0h_18",
"WT6h_CpG_10", "WT6h_CpG_11", "WT6h_CpG_12", "Il4i1ko6h_CpG_25",
"Il4i1ko6h_CpG_26", "Il4i1ko6h_CpG_27", "WT6hns_4", "WT6hns_5",
"WT6hns_6", "Il4i1ko6hns_19", "Il4i1ko6hns_20", "Il4i1ko6hns_21",
"WT24h_CpG_13", "WT24h_CpG_14", "WT24h_CpG_15", "Il4i1ko24h_CpG_28",
"Il4i1ko24h_CpG_29", "Il4i1ko24h_CpG_30", "WT24hns_7", "WT24hns_8",
"WT24hns_9", "Il4i1ko24hns_22", "Il4i1ko24hns_23", "Il4i1ko24hns_24"
), nrows = 36L, elementType = "ANY", elementMetadata = new("DFrame",
rownames = NULL, nrows = 5L, elementType = "ANY", elementMetadata = NULL,
metadata = list(), listData = list(type = c("input", "input",
"input", "input", "input"), description = c("", "", "", "",
""))), metadata = list(), listData = list(id = c(31L, 32L,
33L, 34L, 35L, 36L, 1L, 2L, 3L, 16L, 17L, 18L, 10L, 11L, 12L,
25L, 26L, 27L, 4L, 5L, 6L, 19L, 20L, 21L, 13L, 14L, 15L, 28L,
29L, 30L, 7L, 8L, 9L, 22L, 23L, 24L), sample = c("Il4i1ko_0h_S22",
"Il4i1ko_0h_S21", "Il4i1ko_0h_S20", "WT_0h_S7", "WT_0h_S6", "WT_0h_S5",
"WT0h_1", "WT0h_2", "WT0h_3", "Il4i1ko0h_16", "Il4i1ko0h_17",
"Il4i1ko0h_18", "WT6h_CpG_10", "WT6h_CpG_11", "WT6h_CpG_12",
"Il4i1ko6h_CpG_25", "Il4i1ko6h_CpG_26", "Il4i1ko6h_CpG_27", "WT6hns_4",
"WT6hns_5", "WT6hns_6", "Il4i1ko6hns_19", "Il4i1ko6hns_20", "Il4i1ko6hns_21",
"WT24h_CpG_13", "WT24h_CpG_14", "WT24h_CpG_15", "Il4i1ko24h_CpG_28",
"Il4i1ko24h_CpG_29", "Il4i1ko24h_CpG_30", "WT24hns_7", "WT24hns_8",
"WT24hns_9", "Il4i1ko24hns_22", "Il4i1ko24hns_23", "Il4i1ko24hns_24"
), cell_line = c("Il4i1ko", "Il4i1ko", "Il4i1ko", "WT", "WT",
"WT", "WT", "WT", "WT", "Il4i1ko", "Il4i1ko", "Il4i1ko", "WT",
"WT", "WT", "Il4i1ko", "Il4i1ko", "Il4i1ko", "WT", "WT", "WT",
"Il4i1ko", "Il4i1ko", "Il4i1ko", "WT", "WT", "WT", "Il4i1ko",
"Il4i1ko", "Il4i1ko", "WT", "WT", "WT", "Il4i1ko", "Il4i1ko",
"Il4i1ko"), time_point = c(0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L,
0L, 0L, 0L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 24L,
24L, 24L, 24L, 24L, 24L, 24L, 24L, 24L, 24L, 24L, 24L), treatment = structure(c(2L,
2L, 2L, 2L, 2L, 2L, 1L, 1L, 1L, 1L, 1L, 1L, 2L, 2L, 2L, 2L, 2L,
2L, 1L, 1L, 1L, 1L, 1L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 1L, 1L, 1L,
1L, 1L, 1L), levels = c("none", "CpG"), class = "factor")))


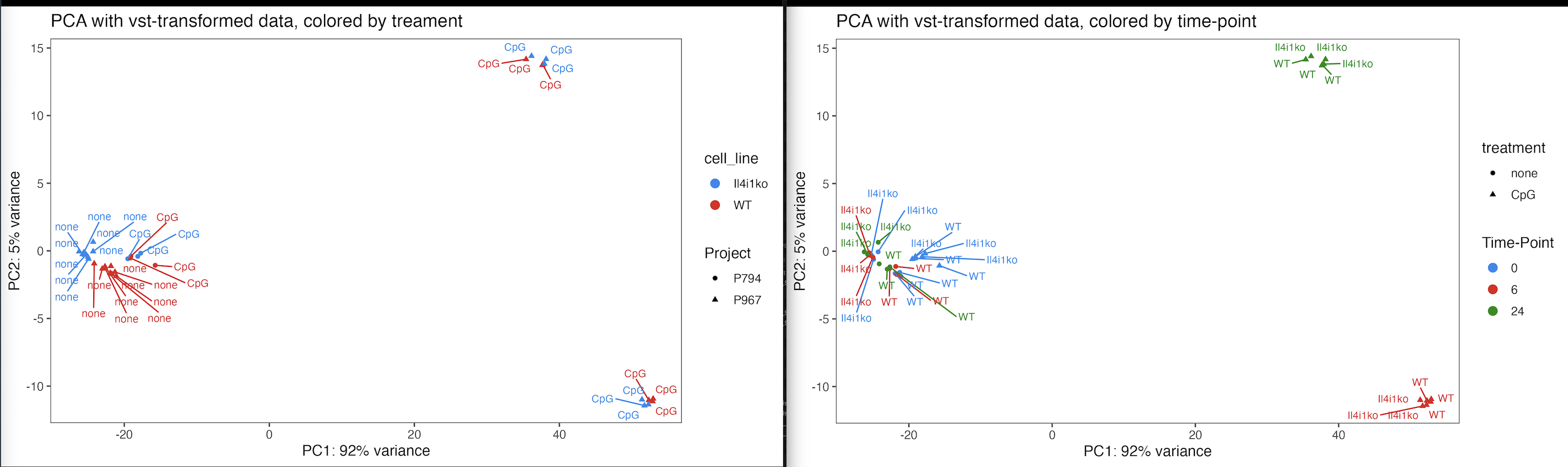
What does your PCA look like? A lot of times, I find that different cell lines are so different from each other, that they should not be directly compared. Your design would be a lot simpler if you split the experiment by cell line.
Thans you swbarnes2 for the answer.
I can see why trying to include all three parameters might be difficult to interpret. But we're interesting in those genes that show a time-dependent differential response to CpG between the KO and the WT (so basically including all three factors). We already did the two analyses splitting the WT and the KO samples, but we would like to try and take it all the way.
I'm attaching here the PCA of the samples to try and show the relationship between them.
Would maybe concatenating the two columns of "cell_line" and "treatment into one make the triple interaction into a two-term interaction and could answer the same question?
thanks
i