I'm trying to understand the results of the AUCell run.
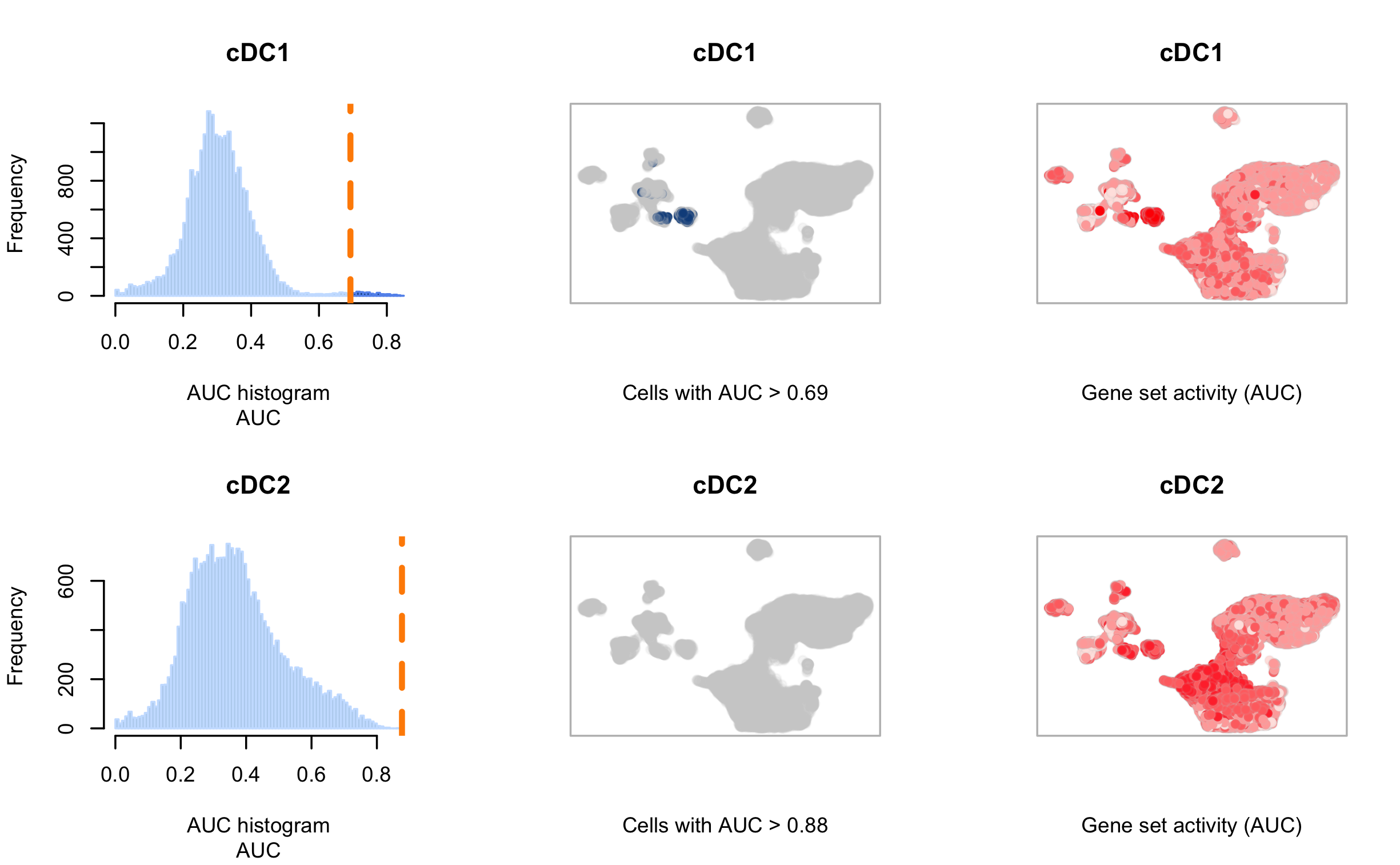
I tested my single-cell data set with specific DC cell-types gene sets. The image below shows two fo the gene sets. for cDC2 i have a very high AUC score, but for some reason no cells are assigned to this category.
If I understand the way the AUC scores are calculated, a high AUC means, many of the genes in the cDC2 gene-set are expressed in the cells, but in the middle plot (created with AUCell_plotTSNE) none of the cells are assigned to this category.
From what I've read, the reason might be connected to the histogram to the left. As I don't have the expected bi-modal behavior, AUCell has difficulties setting a "better" threshold and prefers a somewhat more conservative value. As there are no cells higher of this value, I get an empty UMAP scoring.
On the UMAP to the right though I can see cells with activity in this category. But these cells are not really localized as a cluster, but more spread over multiple regions.
My question is therefore, can I trust AUCell with this value, or do I need to manually assign a lower AUC score to this cell-type. How would I choose a "better" score?
thanks


It sounds like AUCell is struggling with thresholding because your cDC2 gene set doesn t show a clear bimodal distribution. While a high AUC indicates strong gene expression Visit website, AUCell may conservatively assign no cells if the threshold is set too high. In such cases, examining the histogram and considering manual adjustment of the threshold can help. Similarly, the KK33 Game App allows flexibility in tracking progress, letting users adjust goals or difficulty to reflect real performance.