Entering edit mode
Marissa
•
0
@marissa-24334
Last seen 5.2 years ago
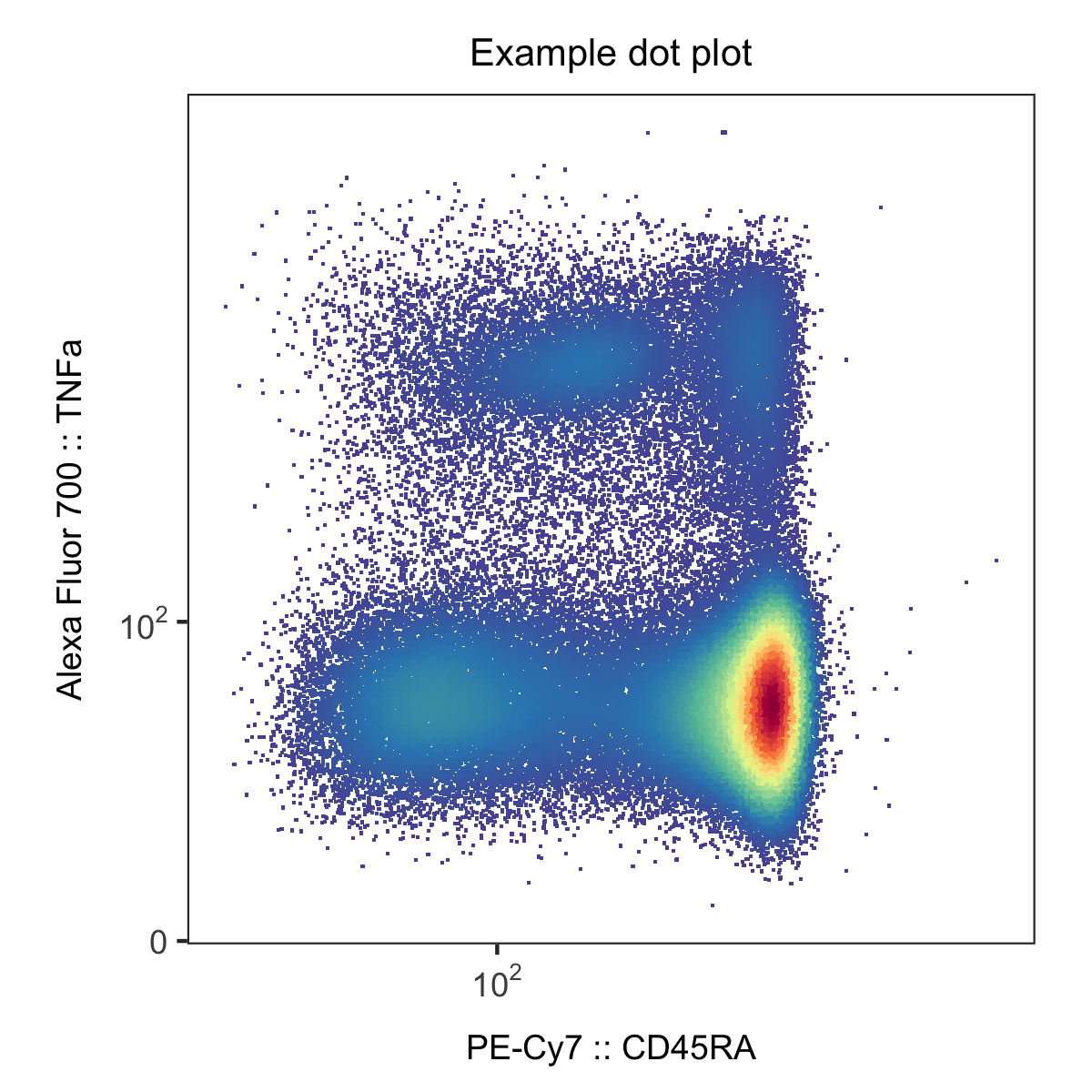
Hi! I am working on developing some flow cytometry dot plots for publication and am a fan of making them using ggcyto. However, I need to add tick marks and a better scale for my current plots because they are sorely lacking under the default setting of scale_x_flowjo_biexp. Does anyone know how to change the limits and breaks and add a nicer scale (say with 10^3, 10^4, etc) to the x and y axes?
library(ggcyto)
library(viridis)
library(CytoML)
library(RColorBrewer)
library(colorspace)
ws <- open_flowjo_xml("Figure 23.wsp")
gs <- flowjo_to_gatingset(ws, name = "R")
plot(gs)
attr(gs, "subset") <- "CD8+"
df <- fortify(gs)
cols <- rev(brewer.pal(10, "Spectral"))
df$d <- densCols(df[,`Comp-PE-Cy7-A`], df[,`Comp-Alexa Fluor 700-A`],
colramp = colorRampPalette(cols))
ggplot(df, aes(x = `Comp-PE-Cy7-A`, y = `Comp-Alexa Fluor 700-A`)) +
geom_point(aes(color = d), shape = '.') +
scale_color_identity() +
theme_mf() +
scale_x_flowjo_biexp() +
scale_y_flowjo_biexp(widthBasis = -100) +
labs(title = "Flow Cytometry Color Dot Plot #1",
subtitle = "Color Palette = 'Reverse Spectral'",
x = "PE-Cy7 :: CD45RA",
y = "Alexa Fluor 700 :: TNFa")


BTW, the data within gs parsed from flowjo_to_gatingset is typically already biexp scale, I wonder if you need/should add extra scale_*_flowjo_biexp layers?