I used DESeq2 to do my Differential gene expression analysis for my RNA seq data. I would like to know what script I can use to generate a manhattan plot where I have the chromosome position on the x axis and p values on the y axis .
Thanks
I used DESeq2 to do my Differential gene expression analysis for my RNA seq data. I would like to know what script I can use to generate a manhattan plot where I have the chromosome position on the x axis and p values on the y axis .
Thanks
You can use this code (just modifying so the -log10 pvalue is plotted instead of the LFC). Also you'd probably not want to bother adding the genes because there will be too many.
~~For more details see the Gviz vignette.~~
This works if you've used tximeta to import the gene expression data. If you've used another pipeline, you'd need to manually add the locations if you haven't already done so.
I'm late answering this question, but leaving this here just in case it useful to anyone.
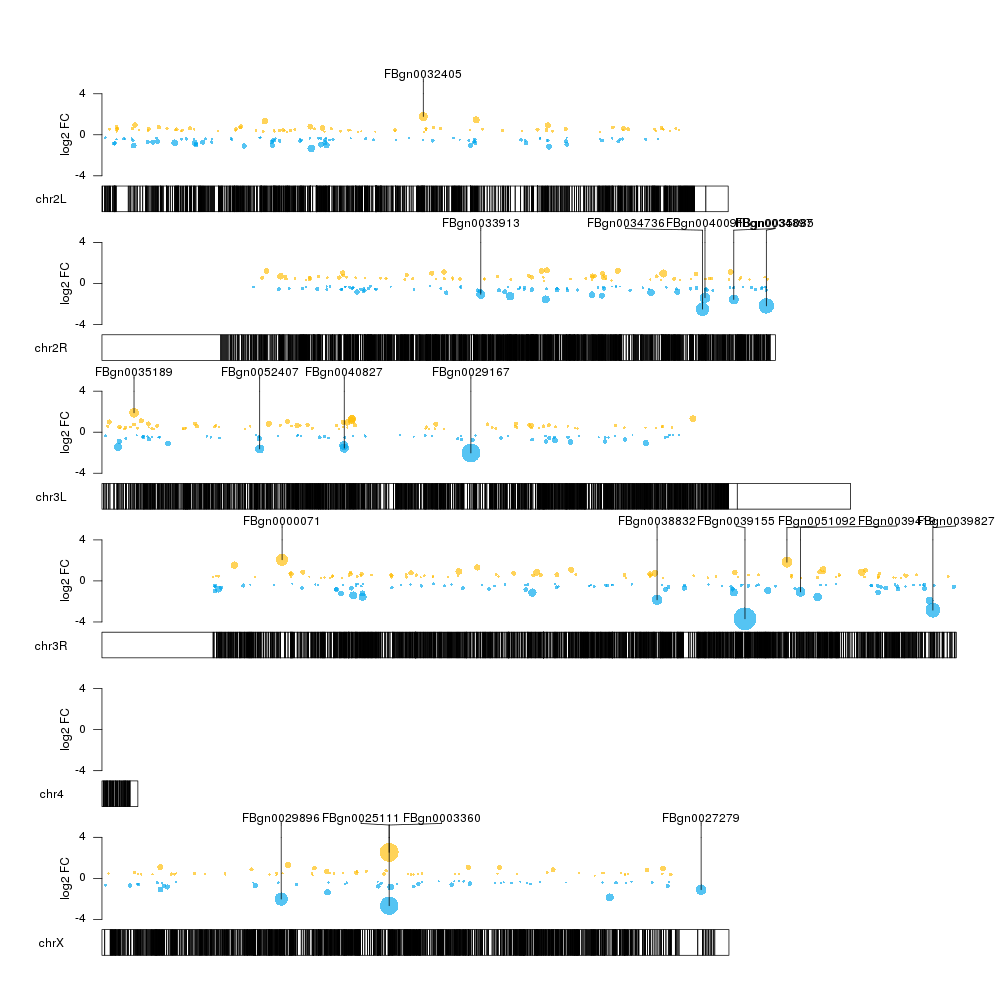
You can create the plot you are asking for with karyoploteR. You could use either a simple kpPoints or make kpPlotManhattan work with you data. In any case, here's an example with DeSeq2 results on drosophila data. In this case y-axis is the fold change and bubble size is the p-value, but you should be able to easily adapt the code to your needs. Oh, and if you want to have all your chromosomes in a single line you'd only need to change a single parameter. You can find more information in the karyoploteR tutorial.
Use of this site constitutes acceptance of our User Agreement and Privacy Policy.
hello Michael thanks for answering my question, I looked at the 1st link you gave , it doesn't have a section on manhattan plots
If you want specific help, can you provide more specific information about what you have?
How have you quantified gene expression? Are you using tximport/tximeta or some other counting tool?
Do you already have a
DESeqDataSetwith chromosome and start position for each gene, or do you need to know also how to obtain this information?I have more specific recommendations after you've obtained the chromosome and start position:
You can use
Sushi::plotManhattanas described here:https://bioconductor.org/packages/release/bioc/vignettes/Sushi/inst/doc/Sushi.pdf
Or you can use
ggbio::plotGrandLinearas described here:http://bioconductor.org/packages/release/bioc/vignettes/ggbio/inst/doc/ggbio.pdf
Hello Michael . I did not use tximport/tximeta. I used featurecounts to get the read counts, I dont have the chromosome positions , how can i include that information ?
Take a look at the Rsubread vignette. They have some information on extracting information about the annotation.