Hello!
I am relatively new to using DESeq2 and was wondering if anyone could help me explain why there is a discrepancy between my normalized counts and the log2FoldChange in the results. Basically, my normalized counts show that a specific gene (dptA and dptB in the example) should be downregulated in my treatment, however, the DESeq results shows a Log2FoldChange which is greater than 0. Below you can find the normalized counts as well as the results from the code below.
coldata <- data.frame(row.names=colnames(countdata), condition)
coldata
dds <- DESeqDataSetFromMatrix(countData=countdata, colData=coldata, design=~condition)
dds
# Run the DESeq2 pipeline
# Get differential expression results
dds <- DESeq(dds)
keep <- rowSums(counts(dds)) >= 10
dds <- dds[keep,]
res<-results(dds,alpha=0.05)
summary(res)
## Write results, Table containing the significantly differential expressed genes
write.table(res,"DESeq2_resuls.txt",sep="\t",col.names=NA)
DESeqResults<-read.table("DESeq2_resuls.txt",sep="\t",header=T)

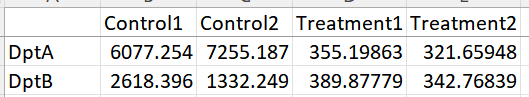
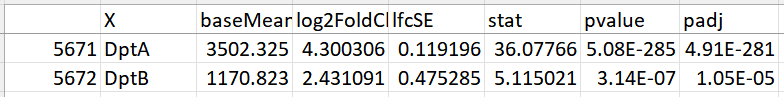

Hello! Thank you so much, turns out the factor levels were being alphabetically ordered changing the order of my treatment and control. GFP being the control and D6 being the treatment got swapped and the program thought that D6 was the control. I fixed it and everything looks correct now.