Hello,
I have a proteomics data set with intensities of the proteins in each individual sample collected during 3 different time points of the experiment: Days 0, 15 and 30. Each subject was sampled repeatedly during each time point; there were 28 subjects in total.
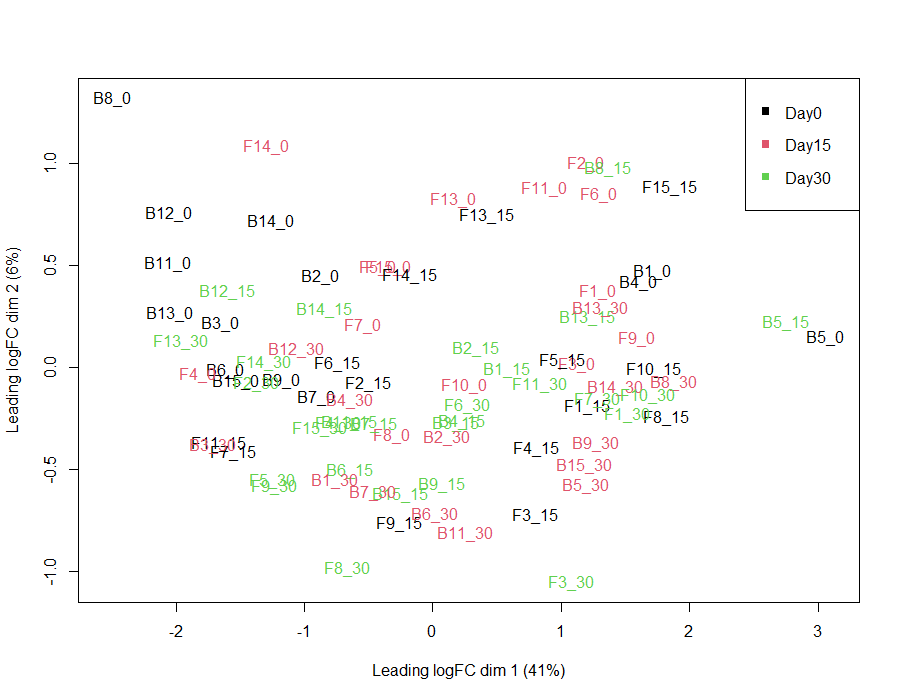
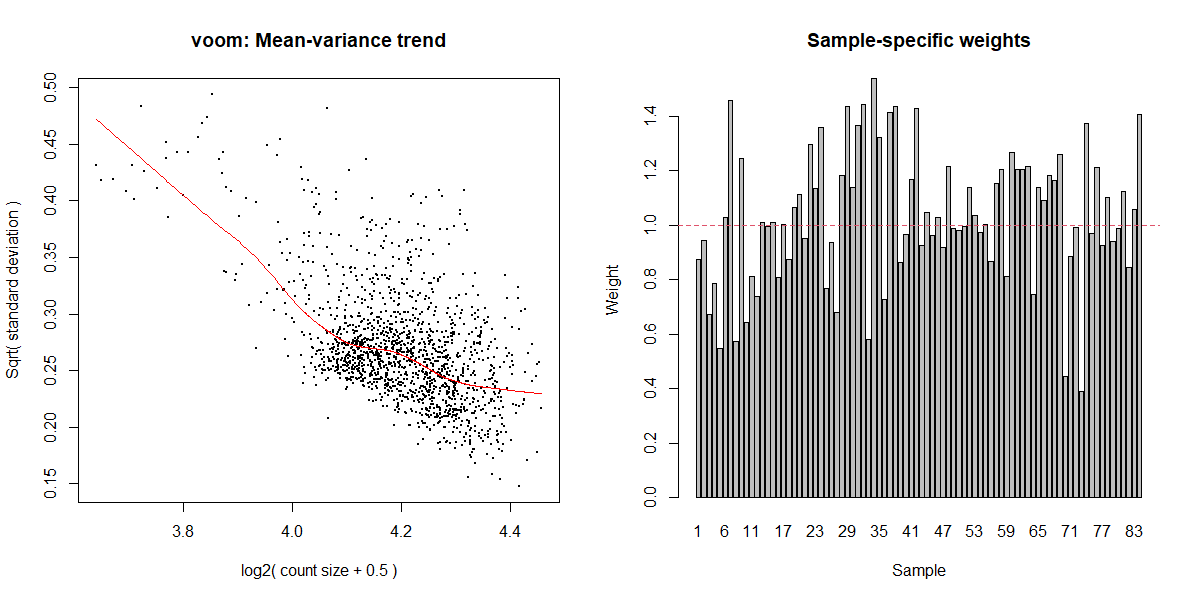
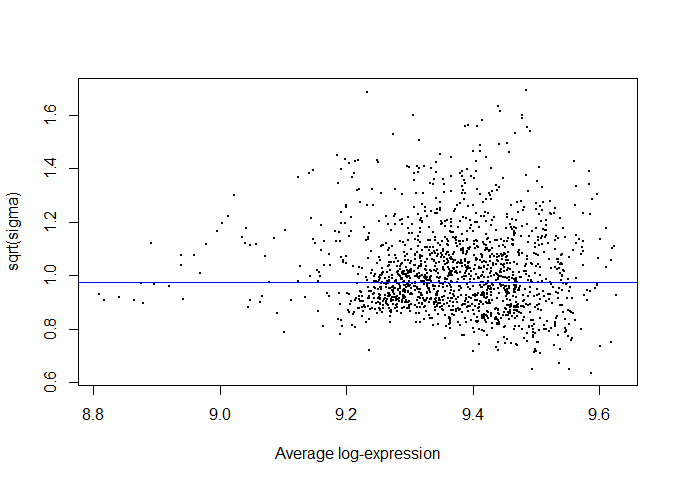
I was just wondering if my limma code could be validated, and if the mean-variance and residual variance plots seem okay? The DE protein lists coincide with what is observed in the MDS plot, however I'm not sure if my mean-variance plot is optimal.
Any feedback on what can be done to correct/improve is greatly appreciated!
#importing protein intensities, row=proteins, columns=SampleID
mc <- read.csv(file.choose(),header = T)
> head(mc)
Sample B1_0 B2_0 B3_0 B4_0 B5_0 B6_0 B7_0 B8_0
1 F3 233359.30 1205534.22 944128.84 1101554.77 128445.59 1240082.66 524326.03 2575986.0
2 ARPC4 40979.78 51904.95 74064.53 38972.67 16760.10 46218.10 60444.73 100100.8
3 KRT17 77261.14 205488.25 819332.98 86556.42 26944.67 509145.45 321247.85 655879.6
4 MRPS36 81783.50 100430.74 129049.67 71059.67 71251.87 313486.79 95901.89 189156.7
5 RBP1 85060.70 381976.65 254617.14 122509.71 80321.20 88639.18 238453.03 5732010.8
6 PRMT1 722479.75 610273.86 419833.22 667370.81 220869.40 1049477.28 688381.30 597083.0
#importing metadata
mc_meta <- read.csv(file.choose(),header = T)
> head(mc_meta) #time "0"=Day0, "1"=Day15, "2"=Day30
Sample Time Subject Day_replicate
1 B1_0 0 S1 0_1
2 B2_0 0 S2 0_1
3 B3_0 0 S3 0_1
4 B4_0 0 S4 0_1
5 B5_0 0 S5 0_1
6 B6_0 0 S6 0_1
mc_meta1 <- mc_meta %>% mutate_if(is.character, as.factor)%>%mutate_if(is.integer, as.factor)
str(mc_meta1)
'data.frame': 84 obs. of 4 variables:
$ Sample : Factor w/ 84 levels "B1_0","B1_15",..: 1 19 22 25 28 31 34 37 40 4 ...
$ Time : Factor w/ 3 levels "0","1","2": 1 1 1 1 1 1 1 1 1 1 ...
$ Subject : Factor w/ 28 levels "S1","S11","S12",..: 1 11 21 23 24 25 26 27 28 2 ...
$ Day_replicate: Factor w/ 6 levels "0_1","0_2","15_1",..: 1 1 1 1 1 1 1 1 1 1 ...
mc_mat <- mc %>% remove_rownames() %>% column_to_rownames("Sample")
mc_mat2<-log2(mc_mat)
subjects <- mc_meta1$Subject
Time1 <- mc_meta1$Time
plotMDS(mc_mat2, col=as.numeric(Time1))
legend("topright", legend=c("Day0", "Day15", "Day30"), col=1:3, pch=15)
mc_mat3<-DGEList(mc_mat2)
mc_norm1<-calcNormFactors(mc_mat3)
design_mc<-model.matrix(~0+Time1)
#estimating correlation between repeated fish measurements
corfit1 <- duplicateCorrelation(mc_mat2, design_mc, block = subjects)
corfit1$consensus.correlation
[1] 0.01975354
> colnames(design_mc)
[1] "Time10" "Time11" "Time12"
aw_mc<-arrayWeights(mc_mat2,design_mc)
contrasts<-makeContrasts(Day15vsDay0=Time11-Time10,
Day30vsDay0=Time12-Time10,
Day30vsDay15=Time12-Time11,
levels=colnames(design_mc))
v <- voomWithQualityWeights(mc_norm1, design_mc, plot=TRUE)
fit1<-lmFit(v,design_mc, block = subjects, correlation = corfit1$consensus.correlation)
fit2<-contrasts.fit(fit1,contrasts=contrasts)
efit<-eBayes(fit2)
plotSA(efit)
summary(decideTests(efit))
Day0vsDay15 Day0vsDay30 Day15vsDay30
Down 0 0 0
NotSig 1547 1544 1547
Up 0 3 0


Thank you! I will try this out again.
Hello again!
The protein intensities are liquid chromatogram intensities. I tried incorporating your suggestions of using cyclic loess normalization and a fixed model instead. Could this code also please be validated?
(I also want to point out that my previous MDS plot was incorrect, as my metadata sample column did not line up correctly with my protein intensity dataset)