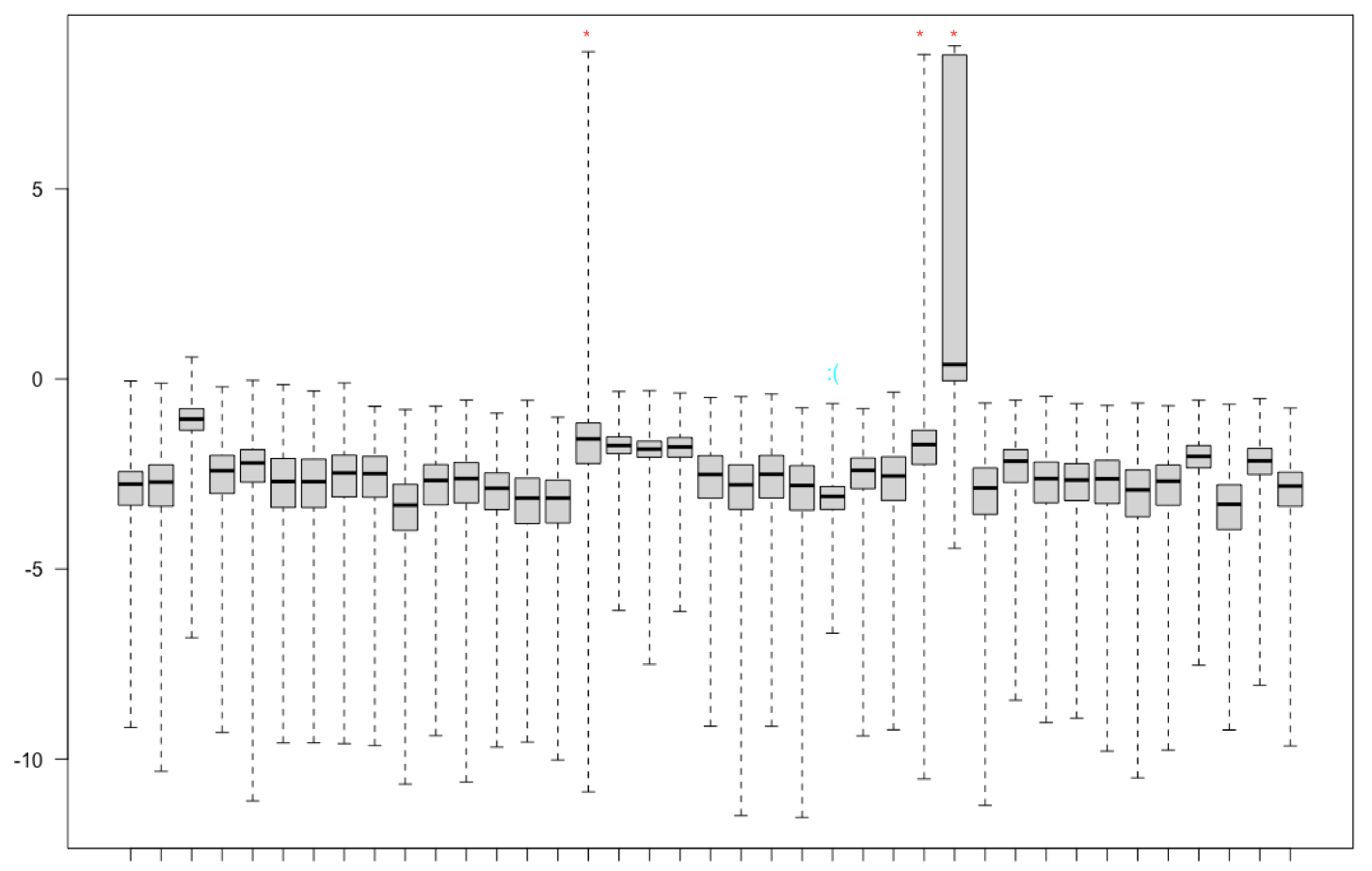
Hi, I recently ran DESeq on my samples and wanted to make sure there were no major outliers in my samples. Here, I tried looking at the dispersion plots, correlation matrix, and Cook's distance.
After plotting Cook's distance, three samples stood out. One had a median distance > 1, but the other two had high variance but normal medians.
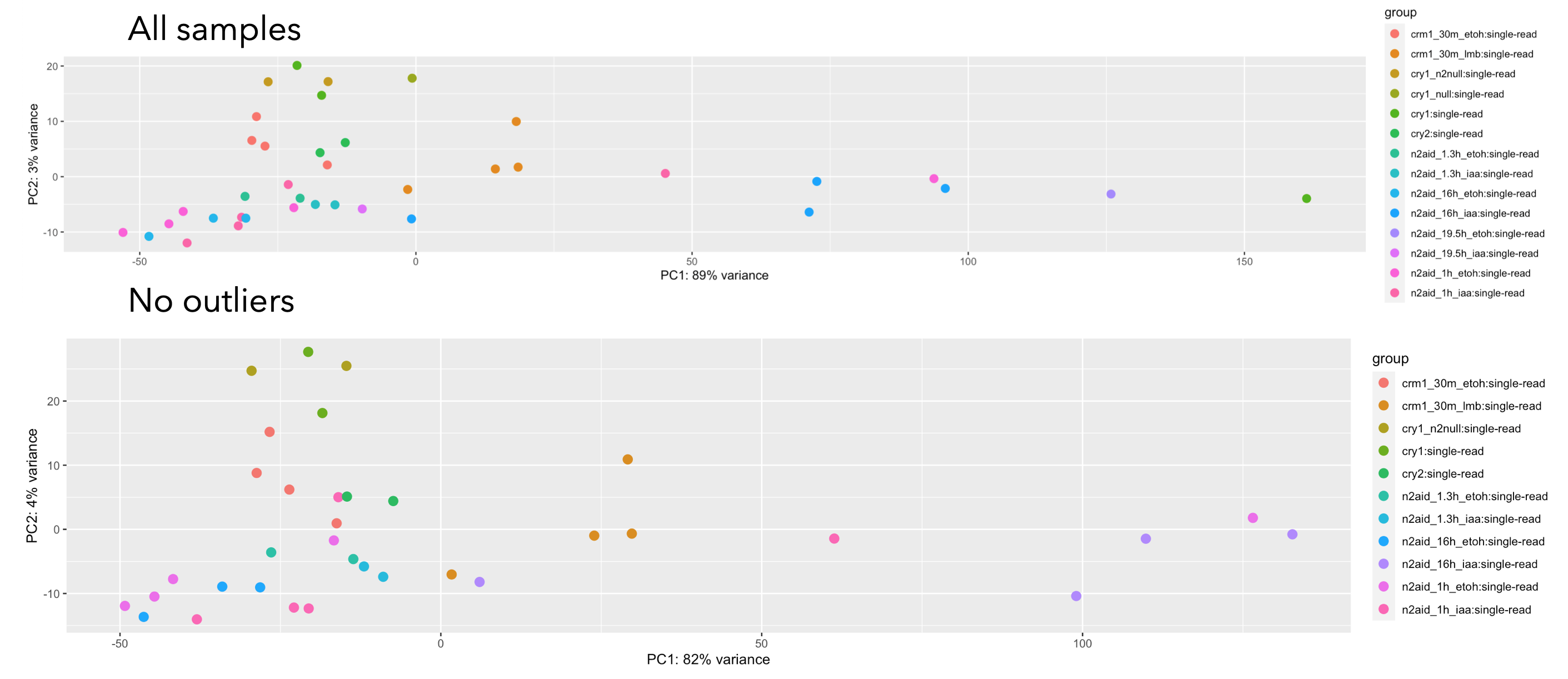
I marked what I thought were outliers in red * but there was a sample that did not correlate well with others marked in cyan. When I remove these four samples from my dataset, my PCA looks like this:
It looks like there's still a lot of variance and I wish I could look at the PCA for one or two samples at a time, but removing the four samples did help the correlation matrix look more clear.
Is this the right way to go about this?