Hello,
I have a large, multi-condition single-cell dataset that I am attempting to use the miloDE package as one part of the analysis.
The object in total has >270,000 cells spanning 5 discrete treatment conditions (15 total samples). For what it is worth, I am expecting the cells used here to represent one overall cell type, so it is a fairly homogenous population.
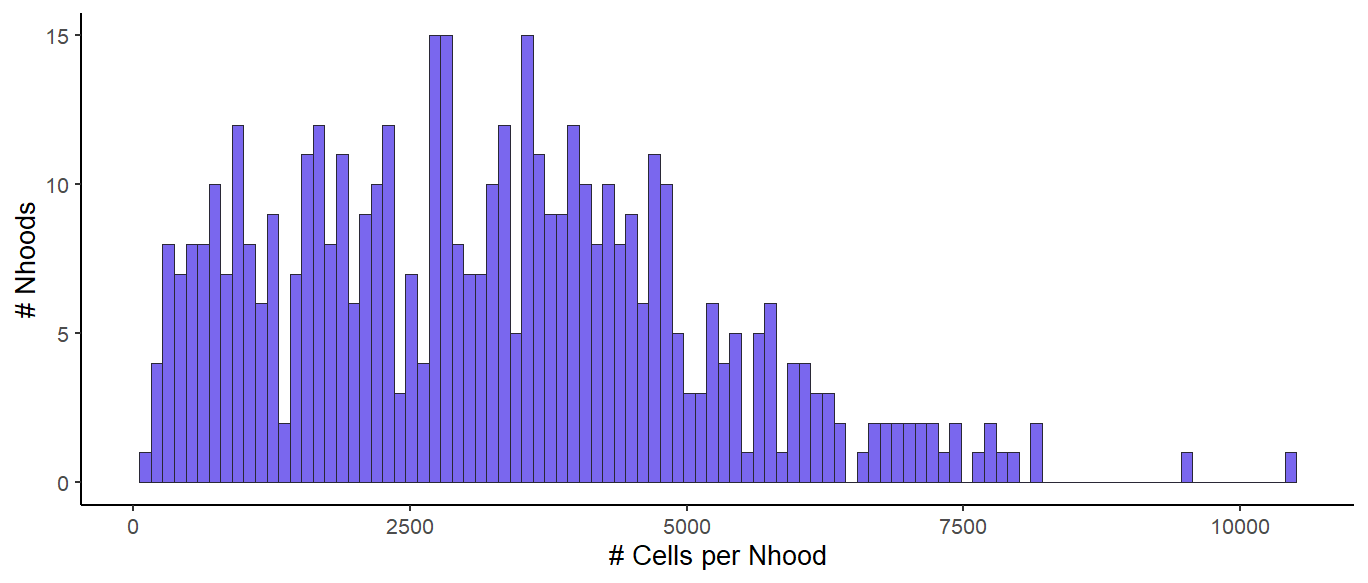
Using a k = 27 for kNN construction, this resulted in 477 neighborhoods with an average size of a few thousand cells per neighborhood (histogram below).
My question is that I have attempted to run differential expression using the "de_test_neighborhoods" function, running just one statistical comparison at a time, but it has taken an extremely long time for the function to conclude. It seems to get 'stuck' on the p-value correction across Nhoods step, where it has sat computing this for tens of hours, even when running it on an HPC cluster. And as of now it has not concluded executing the code or creating the de_stat object.
I was wondering if this is to be expected for such a large dataset as what I have, or if there is perhaps some error in computing that could be happening. Also if anyone has advice on how I might downsample to use fewer cells while keeping rigor across multiple conditions, to streamline this, that would be great too! Thank you!


Is a method that is so computationally expensive really beneficial and necessary? You seem to have biological replication, don't you, so pseudobulk aggregation would be possible. Pseudobulk has been shown to outperform single-cell DE in many setups, there is literature on that. There is also literature that specialiued single-cell methods rarely (if at all) outperform methods originally developed for bulk analysis, but run orders of magnitudes faster. MAybe consider changing method for such an extensive dataset.