Entering edit mode
ecg1g15
▴
30
@ecg1g15-19970
Last seen 5.1 years ago
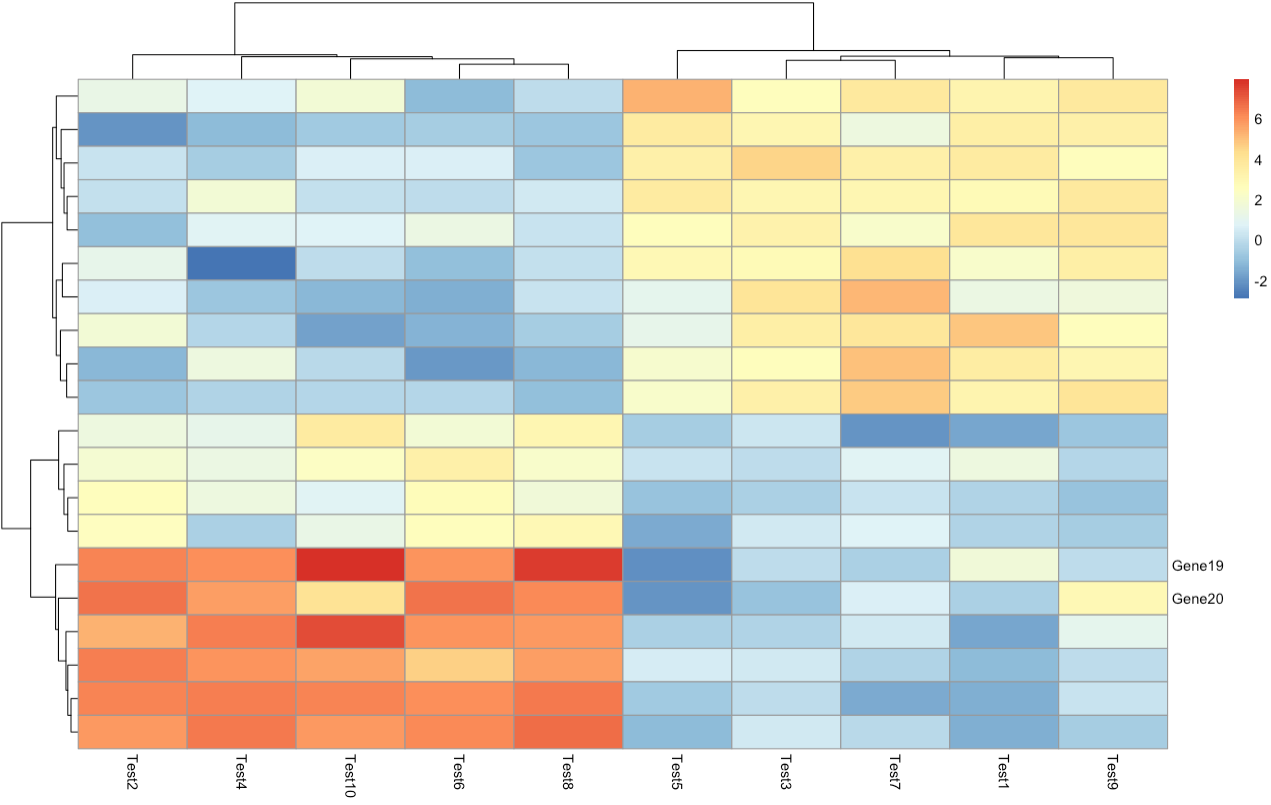
Could you add selected row names to a pheatmap instead of including them all using show_rownames = T?
Something similar was asked few years ago here https://www.biostars.org/p/258462/ for Pheatmap.2 I wonder if now there is any function or package that can allow it other than doing it manually?
I tried something like this but doesn't work
x = matrix(1:16, nrow = 4,
dimnames = list(c("banana","lime","carrot","tomato"), c("A1","A2","B1","B2")))
labgenes<- c(row.names(x)[1], rep('', length(row.names(x))-1))
pheatmap(x, annotation_col = anno, fontsize_row = 10, show_colnames = F, show_rownames = labgenes, color = colorRampPalette(rev(brewer.pal(n = 7, name ="RdYlBu")))(100))


I removed "DESeq2" tag here. Note that tagging a package emails the developers.