Hello everybody,
I have recently started using BioConductor and its flow packages in particular and now would like to learn openCyto. I have a very simple four-color panel for testing purposes. For one of my gating steps, I would like to use the quadGate.tmix method, since the populations are bleeding into each other a little. During my odyssey through the farthest reaches of the internet, I stumpled upon this issue on github: https://github.com/RGLab/flowWorkspace/issues/175
The image on there shows pretty much exactly what I want to achieve, but unfortunately, I have a hard time figuring out how to use the command to actually produce a filter object I can use. Running the quadGate.tmix function with usePrior="no" returns "A list of 4 filters applied to a flowFrame.", which, as far as I understand, contains the four polygon gates defining the quadrants. How do I succesfully apply this to a flowFrame, and how can I visualize it? I can kind of get it to work if I use as filter the first element of the resulting list:
xyplot(`Pacific Blue-A` ~ `PE-A`, testframe, filter=testfilter[[1]])
This results in the following: 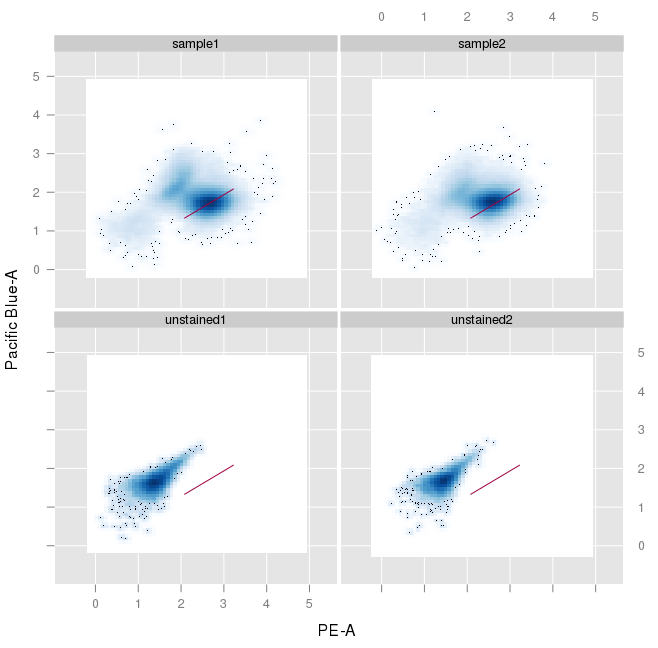
How do I get that to display the full quadrants to me? Also, the thresholding is obviously off, presumably because I ran the filter without any prior. I could not really find any info on that. Maybe I did not look hard enough, but I would much appreciate it if you could point me towards useful resources.
On a different note, I alternatively tried an approach using flowClust. In the openCyto gating template, I set collapseDataForGating to TRUE for this step and was under the impression that this would result in the same gate for all samples. The actual output, however, looks like this:
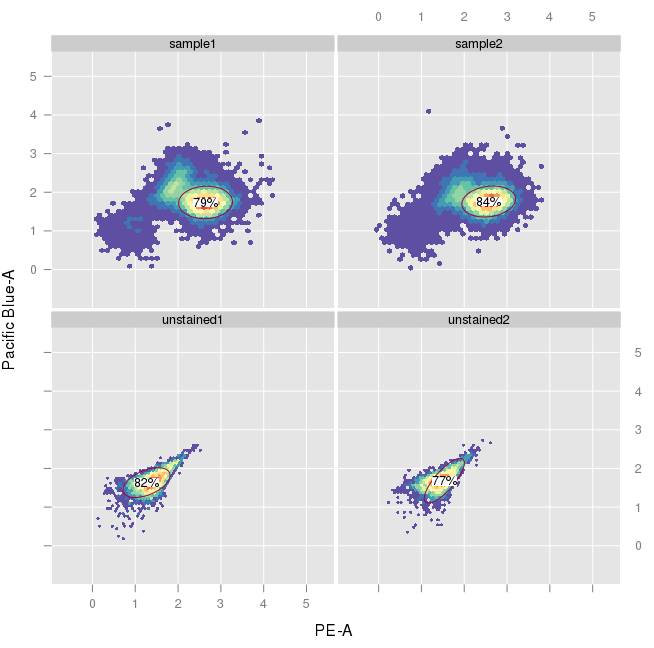
Is there a way to generate a common gate?
I thank you in advance for any help you can provide. I am aware that these are not very specialised questions and that I am probably missing some fundamental concepts here, still I would very much appreciate some useful pointer.
Best,
VT


Turns out to be a bug in `quadGate.tmix` function, I've pushed the fix to openCyto 1.8.3.
As shown in http://rpubs.com/wjiang2/132387, You should set `K` appropriately to achieve the best results.
Note, If you want to speed up flowClust, you can pass a smaller `B` value to 'quadGate.mix`. (default is 500). But you will have to tweak it to see whether it affects the outcome,
Brilliant, thank you! Your help is highly appreciated. I am still at the very beginning of getting to terms with the flow packages, but I hope to use them to their full potential soon. Thanks for your work.