When I use DESeq code as below
sampleFiles <- grep("counts", list.files(directory),value=T) condition <- c("0W","1W","2W","3W","0W","1W","2W","3W","0W","1W","2W","3W") sampleTable <- data.frame(sampleName = sampleFiles, fileName = sampleFiles, condition = condition) sampleTable$condition <- factor(sampleTable$condition) ddsHTSeq <- DESeqDataSetFromHTSeqCount(sampleTable = sampleTable, directory = directory, design= ~ condition)
dds <- DESeq(ddsHTSeq) resultsNames(dds) res <- results(dds, alpha = 0.05, name = "condition_1W_vs_0W", lfcThreshold =1 )
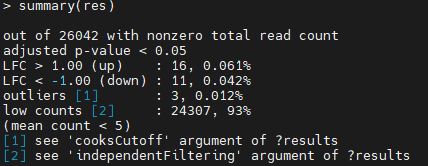
I got a result like attaced picture
I got a 16 up-regulated gene from DESeq2 result.
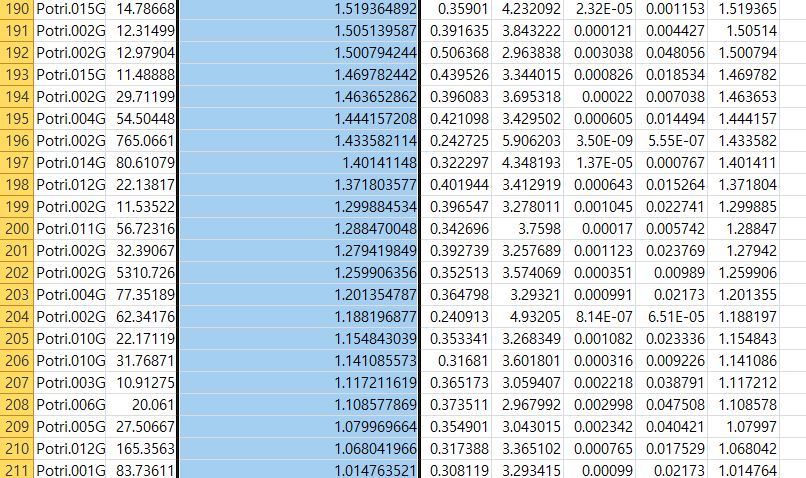
However, when I count number of upregulated gene using excel with same threshold (paj<0.05, logfoldchange >1) I got a 211 up-regulated gene.
Do you know why the result of deg is different despite I used same thereshold?