ddsMat <- DESeqDataSetFromMatrix(countData = fcts,colData = sampleinfo,design = ~ Treatment)
dds <- DESeq(ddsMat,betaPrior=TRUE)
res <- results(dds) # Extract results
library(goseq)
genes=as.integer(p.adjust(res$pvalue[res$log2FoldChange!=0],
method="BH")<.05)
names(genes)=row.names(res[res$log2FoldChange!=0,])
table(genes)
library(TxDb.Hsapiens.UCSC.hg38.knownGene)
TxDb= TxDb.Hsapiens.UCSC.hg38.knownGene
head(supportedOrganisms())
supportedOrganisms()[supportedOrganisms()$Genome=="hg38",]
pwf=nullp(genes,"hg38","ensGene")
head(pwf)
Pathway analysis and GO terms analysis with goseq
0
Entering edit mode
2
Entering edit mode
The error message you received is intended to be self-explanatory, so let's recap. The error says, if I restate (since you included a png to show the output instead of copying and pasting, as requested in the FAQ), 'you gave me a bunch of gene IDs that look like this: ENSG00000227232.5, and yet what I expected looked like this: ENSG00000227232'
And if you look closely at the example IDs, the only difference is a subscript that is a period, followed by a number. Which indicates to me that the answer is to strip off the offending subscript that is being complained about? If you are confused as to how one does that, see ?gsub.
Similar Posts
Loading Similar Posts
Traffic: 990 users visited in the last hour
Use of this site constitutes acceptance of our User Agreement and Privacy Policy.

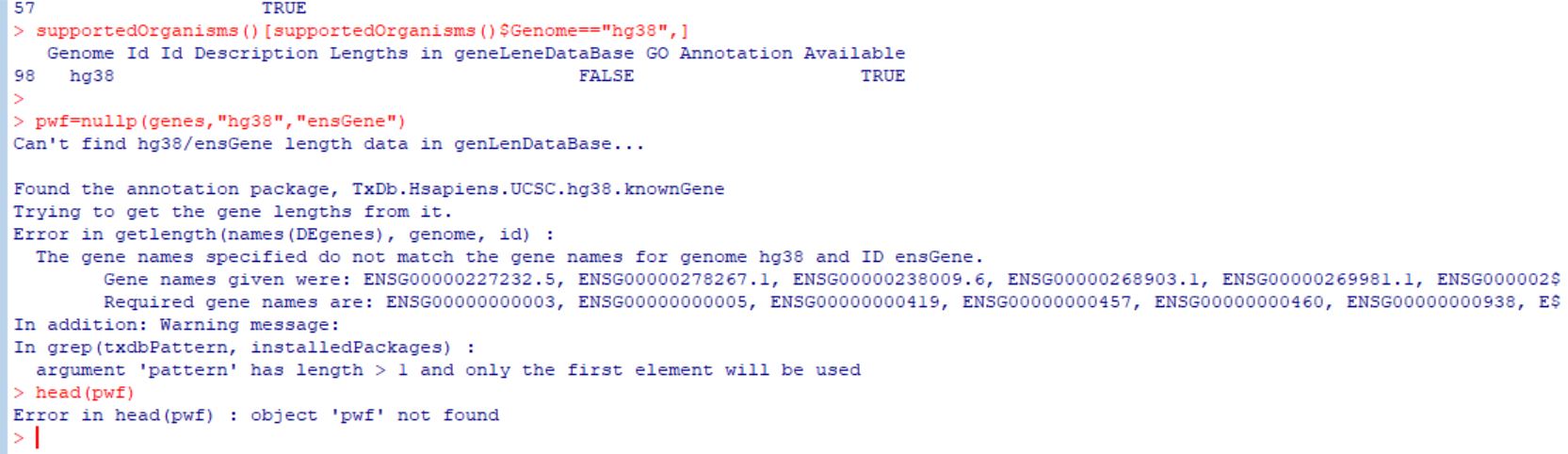

Please, I am stuck with this task. I am trying to perform Gene ontology and KEGG pathway analysis with goseq package. The error message says the gene names specified do not match the gene names for genome hg38 and ID ensGene. I would be glad with any help in achieving this analysis. Thanks in advance