Entering edit mode
I have been unable to run code that has worked in the past, both in the GRaNIE R package and in biomaRT, and it looks like the Ensembl site is unresponsive. I have tried today and last week on 3/20/2025.
Code should be placed in three backticks as shown below
grn <- addTFBS(grn, source = "JASPAR2022",
JASPAR_useSpecificTaxGroup = "vertebrates",
forceRerun = TRUE)
#Error:
#INFO [2025-03-24 10:54:36] Querying JASPAR2022 database. This may take a while.
#INFO [2025-03-24 10:54:42] Retrieving gene annotation for JASPAR TFs. This may take #a while.
#WARN [2025-03-24 10:54:42] Attempt 1 out of 40 failed. Another automatic attempt will be #performed using a different mirror. The error message was: Error: 'curl_parse_url' is not #an exported object from 'namespace:curl'
#Attempts 2-40 here......
#WARN [2025-03-24 10:55:00] Attempt 41 out of 40 failed. Another automatic attempt will #be performed using a different mirror. The error message was: Error: 'curl_parse_url' is #not an exported object from 'namespace:curl'
#ERROR [2025-03-24 10:55:00] A temporary error occured with biomaRt::getBM or #biomaRt::useEnsembl despite trying 40 times via different mirrors. This is often caused by #an unresponsive Ensembl site. Try again at a later time. Note that this error is not caused #by GRaNIE but external services.
# An error occurred. See details above. If you think this is a bug, please contact us. #
#Error in .checkAndLogWarningsAndErrors(NULL, error_Biomart, isWarning = FALSE) :
# A temporary error occured with biomaRt::getBM or biomaRt::useEnsembl despite trying #40 times via different mirrors. This is often caused by an unresponsive Ensembl site. Try #again at a later time. Note that this error is not caused by GRaNIE but external services.
Here's another example of simple code that has worked in the past:
listEnsembl()
listEnsemblGenomes()
#Error: 'curl_parse_url' is not an exported object from 'namespace:curl'
sessionInfo( )
R version 4.4.1 (2024-06-14 ucrt)
Platform: x86_64-w64-mingw32/x64
Running under: Windows 11 x64 (build 22621)
Matrix products: default
locale:
[1] LC_COLLATE=English_United States.utf8 LC_CTYPE=English_United States.utf8 LC_MONETARY=English_United States.utf8
[4] LC_NUMERIC=C LC_TIME=English_United States.utf8
time zone: America/New_York
tzcode source: internal
attached base packages:
[1] parallel grid compiler stats4 tools stats graphics grDevices utils datasets methods base
other attached packages:
[1] VennDiagram_1.7.3 futile.logger_1.4.3 UpSetR_1.4.0
[4] survival_3.8-3 rpart_4.1.24 mgcv_1.9-1
[7] MASS_7.3-64 foreign_0.8-88 cluster_2.1.8
[10] boot_1.3-31 TxDb.Hsapiens.UCSC.hg19.knownGene_3.2.2 preprocessCore_1.68.0
[13] knitr_1.49 zlibbioc_1.52.0 vctrs_0.6.5
[16] utf8_1.2.4 lubridate_1.9.4 forcats_1.0.0
[19] readr_2.1.5 tidyverse_2.0.0 tidyselect_1.2.1
[22] tidyr_1.3.1 tibble_3.2.1 summarytools_1.0.1
[25] stringr_1.5.1 stringi_1.8.4 scales_1.3.0
[28] S4Arrays_1.6.0 rstudioapi_0.17.1 RSQLite_2.3.9
[31] rlang_1.1.5 remotes_2.5.0 reactome.db_1.89.0
[34] RCurl_1.98-1.16 Rcpp_1.0.14 R6_2.5.1
[37] purrr_1.0.4 png_0.1-8 pkgconfig_2.0.3
[40] pheatmap_1.0.12 pillar_1.10.1 NoRCE_1.18.0
[43] nlme_3.1-167 munsell_0.5.1 msigdbr_7.5.1
[46] memoise_2.0.1 magrittr_2.0.3 locfit_1.5-9.11
[49] lifecycle_1.0.4 lattice_0.22-6 KEGGREST_1.46.0
[52] impute_1.80.0 httr_1.4.7 gtable_0.3.6
[55] gridExtra_2.3 GO.db_3.20.0 glue_1.8.0
[58] ggrepel_0.9.6 GenomeInfoDbData_1.2.13 generics_0.1.3
[61] fgsea_1.32.2 fastmatch_1.1-6 fastmap_1.2.0
[64] fansi_1.0.6 ellipsis_0.3.2 dplyr_1.1.4
[67] plyr_1.8.9 devtools_2.4.5 usethis_3.1.0
[70] DESeq2_1.46.0 DBI_1.2.3 crayon_1.5.3
[73] cowplot_1.1.3 colorspace_2.1-1 car_3.1-3
[76] carData_3.0-5 cachem_1.1.0 blob_1.2.4
[79] bitops_1.0-9 bit_4.5.0.1 babelgene_22.9
[82] assertthat_0.2.1 abind_1.4-8 curl_5.2.3
[85] BiocVersion_3.20.0 IHW_1.34.0 EDASeq_2.40.0
[88] ShortRead_1.64.0 GenomicAlignments_1.42.0 Rsamtools_2.22.0
[91] ReactomePA_1.50.0 variancePartition_1.36.3 ggplot2_3.5.1
[94] BiocParallel_1.40.0 clusterProfiler_4.14.4 DOSE_4.0.0
[97] WGCNA_1.73 fastcluster_1.2.6 dynamicTreeCut_1.63-1
[100] rbioapi_0.8.2 motifmatchr_1.28.0 TFBSTools_1.44.0
[103] JASPAR2022_0.99.8 BiocFileCache_2.14.0 dbplyr_2.5.0
[106] TxDb.Rnorvegicus.UCSC.rn6.refGene_3.4.6 org.Rn.eg.db_3.20.0 ChIPseeker_1.42.1
[109] ensembldb_2.30.0 AnnotationFilter_1.30.0 GenomicFeatures_1.58.0
[112] AnnotationDbi_1.68.0 BSgenome.Rnorvegicus.UCSC.rn6_1.4.1 BSgenome_1.74.0
[115] rtracklayer_1.66.0 BiocIO_1.16.0 Biostrings_2.74.1
[118] XVector_0.46.0 BiocManager_1.30.25 csaw_1.40.0
[121] SummarizedExperiment_1.36.0 Biobase_2.66.0 MatrixGenerics_1.18.1
[124] matrixStats_1.5.0 GenomicRanges_1.58.0 GenomeInfoDb_1.42.3
[127] IRanges_2.40.1 S4Vectors_0.44.0 BiocGenerics_0.52.0
[130] biomaRt_2.62.1 Matrix_1.7-2 data.table_1.16.4
[133] edgeR_4.4.2 limma_3.62.2 GRaNIE_1.10.0
loaded via a namespace (and not attached):
[1] R.methodsS3_1.8.2 progress_1.2.3 urlchecker_1.0.1 nnet_7.3-20
[5] poweRlaw_1.0.0 ggtangle_0.0.6 digest_0.6.37 corpcor_1.6.10
[9] shape_1.4.6.1 deldir_2.0-4 magick_2.8.5 reshape2_1.4.4
[13] httpuv_1.6.15 foreach_1.5.2 qvalue_2.38.0 withr_3.0.2
[17] xfun_0.50 ggfun_0.1.8 gson_0.1.0 profvis_0.4.0
[21] tidytree_0.4.6 GlobalOptions_0.1.2 gtools_3.9.5 R.oo_1.27.0
[25] Formula_1.2-5 prettyunits_1.2.0 promises_1.3.2 restfulr_0.0.15
[29] miniUI_0.1.1.1 UCSC.utils_1.2.0 base64enc_0.1-3 ggraph_2.2.1
[33] polyclip_1.10-7 SparseArray_1.6.1 xtable_1.8-4 doParallel_1.0.17
[37] evaluate_1.0.3 hms_1.1.3 filelock_1.0.3 later_1.4.1
[41] viridis_0.6.5 ggtree_3.14.0 XML_3.99-0.18 Hmisc_5.2-2
[45] iterators_1.0.14 pwalign_1.2.0 caTools_1.18.3 minqa_1.2.8
[49] gridGraphics_0.5-1 graphlayouts_1.2.2 rapportools_1.1 codetools_0.2-20
[53] slam_0.1-55 remaCor_0.0.18 mime_0.12 splines_4.4.1
[57] circlize_0.4.16 interp_1.1-6 seqLogo_1.72.0 lme4_1.1-36
[61] fs_1.6.5 checkmate_2.3.2 Rdpack_2.6.2 pkgbuild_1.4.6
[65] ggplotify_0.1.2 statmod_1.5.0 tzdb_0.4.0 fANCOVA_0.6-1
[69] tweenr_2.0.3 RhpcBLASctl_0.23-42 rbibutils_2.3 viridisLite_0.4.2
[73] numDeriv_2016.8-1.1 graphite_1.52.0 rmarkdown_2.29 broom_1.0.7
[77] AnnotationHub_3.14.0 patchwork_1.3.0 graph_1.84.1 farver_2.1.2
[81] reformulas_0.4.0 aod_1.3.3 tidygraph_1.3.1 yaml_2.3.10
[85] latticeExtra_0.6-30 cli_3.6.3 mvtnorm_1.3-3 sessioninfo_1.2.3
[89] lambda.r_1.2.4 backports_1.5.0 annotate_1.84.0 timechange_0.3.0
[93] rjson_0.2.23 ape_5.8-1 jsonlite_1.8.9 bit64_4.6.0-1
[97] yulab.utils_0.2.0 CNEr_1.42.0 futile.options_1.0.1 metapod_1.14.0
[101] GOSemSim_2.32.0 R.utils_2.12.3 pbkrtest_0.5.3 lazyeval_0.2.2
[105] pander_0.6.5 shiny_1.10.0 htmltools_0.5.8.1 enrichplot_1.26.6
[109] rappdirs_0.3.3 formatR_1.14 tcltk_4.4.1 TFMPvalue_0.0.9
[113] httr2_1.1.0 treeio_1.30.0 jpeg_0.1-10 EnvStats_3.0.0
[117] igraph_2.1.4 gplots_3.2.0 fdrtool_1.2.18 pkgload_1.4.0
[121] aplot_0.2.4 nloptr_2.1.1 DirichletMultinomial_1.48.0 DelayedArray_0.32.0
[125] plotrix_3.8-4 ProtGenerics_1.38.0 htmlTable_2.4.3 ggforce_0.4.2
[129] xml2_1.3.6 KernSmooth_2.23-26 pryr_0.1.6 htmlwidgets_1.6.4
[133] aroma.light_3.36.0 RColorBrewer_1.1-3 hwriter_1.3.2.1 lmerTest_3.1-3
[137] lpsymphony_1.34.0


Can you try updating the curl package to the latest version. The
curl_parse_url()function was only added in version 6.0 and you're using 5.2.3. You can do this via:Then test the code again.
I am also having issues using BioMart to run code that has worked before. The problem first occurred last week. As I have encountered these problems before, I patiently did something else for serveral hours while waiting for it to resolve, which it did. Now it has broken again. i can download the mart with useEnsembl() The problem occurs when I use getBM() which results in the following error. Error in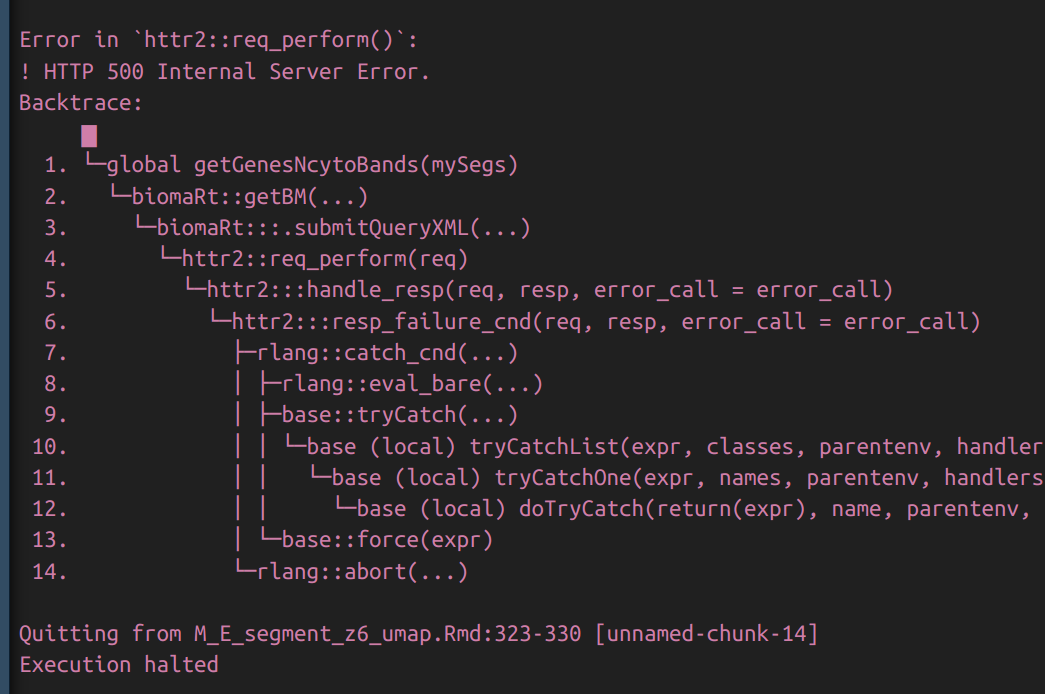 My curl version was already > 6.0
But I have updated anyway to 6.2.2
My curl version was already > 6.0
But I have updated anyway to 6.2.2
httr2::req_perform(): ! HTTP 500 Internal Server Error.R version 4.4.3 (2025-02-28) Platform: x86_64-pc-linux-gnu Running under: Ubuntu 22.04.5 LTS
Matrix products: default BLAS: /usr/lib/x86_64-linux-gnu/blas/libblas.so.3.10.0 LAPACK: /usr/lib/x86_64-linux-gnu/lapack/liblapack.so.3.10.0
locale: 1 LC_CTYPE=en_AU.UTF-8 LC_NUMERIC=C LC_TIME=en_AU.UTF-8 LC_COLLATE=en_AU.UTF-8
[5] LC_MONETARY=en_AU.UTF-8 LC_MESSAGES=en_AU.UTF-8 LC_PAPER=en_AU.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C LC_MEASUREMENT=en_AU.UTF-8 LC_IDENTIFICATION=C
time zone: Australia/Sydney tzcode source: system (glibc)
attached base packages: 1 stats graphics grDevices utils datasets methods base
other attached packages: 1 Biobase_2.66.0 BiocGenerics_0.52.0
loaded via a namespace (and not attached): 1 tidyselect_1.2.1 gridBase_0.4-7 viridisLite_0.4.2
[4] dplyr_1.1.4 viridis_0.6.5 fastmap_1.2.0
[7] lazyeval_0.2.2 rpart_4.1.24 digest_0.6.37
[10] factoextra_1.0.7 lifecycle_1.0.4 cluster_2.1.8.1
[13] magrittr_2.0.3 compiler_4.4.3 rlang_1.1.5
[16] Hmisc_5.2-3 rngtools_1.5.2 tools_4.4.3
[19] yaml_2.3.10 corrplot_0.95 data.table_1.17.0
[22] NMF_0.28 knitr_1.50 htmlwidgets_1.6.4
[25] plyr_1.8.9 RColorBrewer_1.1-3 registry_0.5-1
[28] foreign_0.8-88 purrr_1.0.4 nnet_7.3-20
[31] grid_4.4.3 stats4_4.4.3 colorspace_2.1-1
[34] ggplot2_3.5.1 scales_1.3.0 iterators_1.0.14
[37] MASS_7.3-65 cli_3.6.4 rmarkdown_2.29
[40] generics_0.1.3 rstudioapi_0.17.1 httr_1.4.7
[43] reshape2_1.4.4 stringr_1.5.1 zlibbioc_1.52.0
[46] parallel_4.4.3 limSolve_1.5.7.1 BiocManager_1.30.25
[49] XVector_0.46.0 base64enc_0.1-3 vctrs_0.6.5
[52] jsonlite_1.9.1 bookdown_0.42 naniar_1.1.0
[55] IRanges_2.40.1 S4Vectors_0.44.0 ggrepel_0.9.6
[58] visdat_0.6.0 htmlTable_2.4.3 Formula_1.2-5
[61] dendextend_1.19.0 foreach_1.5.2 plotly_4.10.4
[64] tidyr_1.3.1 glue_1.8.0 codetools_0.2-20
[67] DT_0.33 stringi_1.8.4 gtable_0.3.6
[70] GenomeInfoDb_1.42.3 GenomicRanges_1.58.0 UCSC.utils_1.2.0
[73] quadprog_1.5-8 munsell_0.5.1 tibble_3.2.1
[76] pillar_1.10.1 htmltools_0.5.8.1 GenomeInfoDbData_1.2.13
[79] R6_2.6.1 CINSignatureQuantification_1.2.0 doParallel_1.0.17
[82] evaluate_1.0.3 lpSolve_5.6.23 backports_1.5.0
[85] Rcpp_1.0.14 checkmate_2.3.2 gridExtra_2.3
[88] xfun_0.51 pkgconfig_2.0.3
I think this is an unrelated issue. The original post includes the message
Error: 'curl_parse_url' is not an exported object from 'namespace:curl'indicating this is an issue with the curl package.Your error comes from the Ensembl BioMart server, which is becoming increasingly unreliable. Unfortunately neither Biocondutor nor the biomaRt package maintainer have any control over the Ensembl BioMart server. If you'd like to ask about its status, the Ensembl helpdesk can be contacted at https://www.ensembl.org/Help/Contact
Thanks for the link. I will contact them directly, I wish they had some sort of status page.